A special collection to commemorate the Tree of Life’s 150th Anniversary
| 1 December, 2016 | Thomas Ingraham |
|

|

The phylogenetic ‘Tree of Life’, arguably the most elegant representation of the evolutionary process, perhaps of any biological process, is 150 years old this year. To commemorate this anniversary we have created a special collection of phylogenetics articles published on F1000Research.
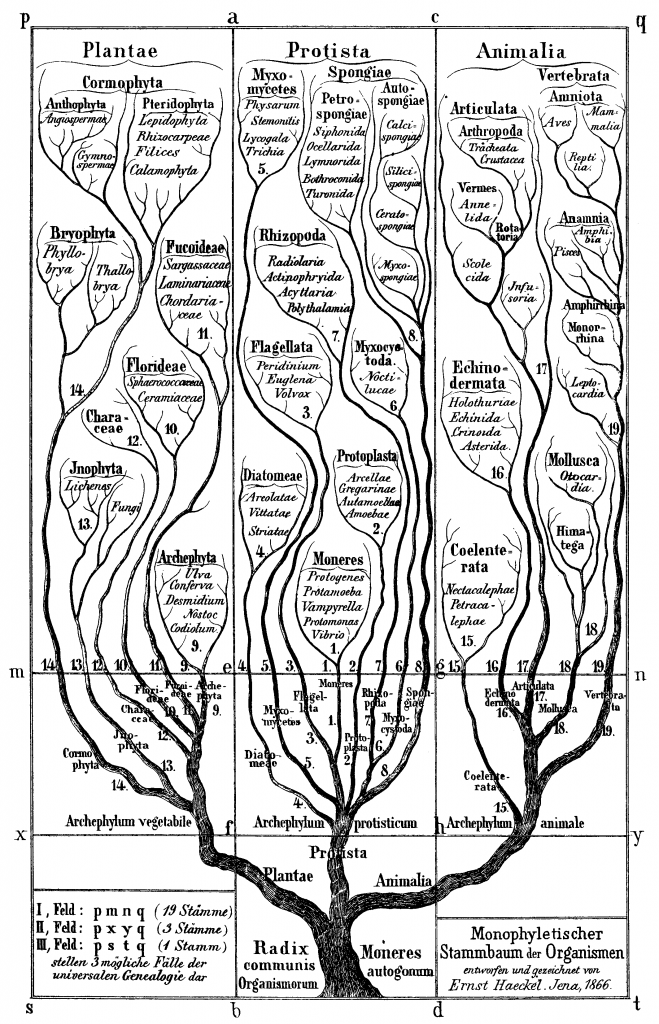
Darwin first sketched an evolutionary tree in an 1837 notebook entry, but both this and the illustration included in On the Origin of Species were abstractions used to illustrate the path evolution takes. It was the artist-cum-scientist Ernst Haeckel who drew the first* evolutionary tree that attempted to represent the actual historical inter-relatedness of all major eukaryotic life forms (pictured) in his 1866 Generelle Morphologie der Organismen. It was also he who coined the term ‘phylogeny (or ‘phylogenie’ in his native German) to describe the evolutionary relationship between species.
Phylogenetics has come a long way in the last century and a half; relationships are based on molecular sequences rather than morphology, tree dimensions are computationally calculated rather than drawn freehand, and some phylogenies have morphed from trees to networks. Along with previous phylogenetic-focused articles published in F1000Research, this progress is well represented by the five latest articles published in the collection:
Luis Arteaga-Figueroa, Valentina Sánchez-Bermúdez and Nicolás Franco-Sierra of Universidad EAFIT updates previous morphology and single gene based phylogenies of Ctenophora (comb jellies) with multi gene analyses. This contribution is particularly apt for this collection considering Haeckel’s special fondness of Ctenophores (pictured), to the extent that an entire family of comb jellies, the Haeckeliidae, are named after him.
Guillaume Bernard, Mark Ragan and Cheong Xin Chan of The University of Queensland use a scalable, dynamic shared k-mers approach to reconstruct the relationship networks between 143 bacterial and archael genomes. Representing these networks dynamically allows biological questions of interest to be formulated and addressed quickly and in a visually intuitive manner.
Will Pearse of Utah State University gives a short review of recent major advances in phylogenetic visualizations, and outlines a new kind of graphic, the fibre plot. This approach uses the metaphor of sections through a tree to describe change in a phylogeny and can be represented as an animation.
Martin Ryberg of Uppsala University presents Phylommand, a new software package pipeline comprised of four programs that can create, manipulate, and/or analyze phylogenetic trees or pairwise alignments.
Alexey Sergeev of the Russian Federal Research Institute of Fisheries and Oceanography deciphers the phylogenetic relationship between Russian, Persian and Siberian sturgeon populations, confirming that despite the genetic isolation of the Persian sturgeon from the Russian sturgeon in the Caspian Sea, the Persian sturgeon is a young species.
You can track the collection (by clicking the alarm clock icon) to be notified of other phylogenetic articles that will be published in coming weeks and beyond.
* This is admittedly contentious as several phylogenetic trees were published in 1866 with St George Mivart, Franz Hilgendorf and Jean Gaudry all having similarly good claims.

|