www.idosr.org
©IDOSR PUBLICATIONS
International Digital Organization for Scientific Research
Aziz et al
ISSN: 2579-0781
IDOSR JOURNAL OF EXPERIMENTAL SCIENCES 8(1) 60-71, 2022. Prevalence of Animal African Trypanosomiasis in Lira District Using Two Selected Diagnostic Methods:Microscopy and ITS-Polymerase Chain Reaction Amplification
Aziz1 , Simon
Peter Musinguzi2 , Martin
Odoki1 and Gift Witto1
1Department of Microbiology and Immunology, Kampala International University, Western Campus,Ishaka-Bushenyi,Uganda.
2DepartmentofAnimalSciences,FacultyofAgricultureandEnvironmentalSciences,Kabale University,Uganda.
ABSTRACT
Trypanosomes are the causative agents of animal African trypanosomiasis (AAT) and human African trypanosomiasis (HAT), the former affecting domestic animals prevalent in sub-Saharan Africa. The main species causing AAT in cattle are T.congolense, T.vivax and T.b.brucei. Northern Uganda has been politically unstable with no form of vector control in place. The return of displaced inhabitants led to the restocking of cattle from AAT endemic areas. It was thus important to estimate the burden of trypanosomiasis in the region.ThisstudywasdesignedtocomparetheprevalenceofanimalAfricantrypanosomes in cattle in Lira District using microscopy and polymerase chain reaction amplification (PCR)methods.Inthiscross-sectionalstudy,atotalof254cattlefromthethreevillagesof Acanakwo A, Barropok and Acungkena in Lira District, Uganda, were selected by simple random sampling technique and screened for trypanosomiasis using microscopy and PCR methods. The prevalence of trypanosomiasis according to microscopic results was 5/254 (2.0%)ascomparedto11/254(4.3%)trypanosomiasisprevalenceaccordingtoPCRanalysis. The prevalence of trypanosomiasis infection in the animal studied is 11/254 (4.3%). Trypanosomacongolense was the most dominant Trypanosomes species with a proportion of 9/11 (81.8%), followed by T.vivax 1/11 (9.1%) and mixed infection of T. congolense/T.vivax1/11 (9.1%). Barropok village had the highest prevalence of trypanosomiasis with 6/11 (54.5%). There is statistical significant relationship (OR=6.041; 95% CI: 1.634-22.331; p<0.05) between abnormal PCV and trypanosome infection. Polymerase reaction amplification was the most reliable diagnostic method due to its high sensitivity and specificity as compared to the conventional microscopic method. Polymerasereactionamplificationappearstohaveadequateaccuracytosubstitutethe use of a microscope where facilities allow. This study, therefore, underscores the urgent need forlocal surveillance schemes more especially atthe grassroots in Uganda to provide data forreferenceguidelinesdevelopmentneededforcontroloftrypanosomiasisinUganda.
Keywords:Trypanosomiasis,PCR,LiraDistrict,Microscopy
INTRODUCTION
Animal African Trypanosomiasis (AAT) is one of the most significant vector-borne diseasesofdomesticanimalsinthetsetse belt of Africa including East Africa [1] AATis causedby Trypanosomaspp which aretransmittedcyclicallybythetsetsefly belonging to the Glossina spp. Animal African Trypanosomiasis (AAT) affects a wide range of hosts namely goats, sheep, donkeys and cattle. The disease is
characterized by the availability of parasites in the blood and alternating fever [2]. Anemia usually develops in infected animals, accompanied by weight loss, body loss, miscarriage, abortion decreased productivity and sometimes mortality [3]. Animal African Trypanosomiasis causes more than 3 million animals to die each year with 50 million animals at risk of infection [4].
www.idosr.org
Therefore, AAT is a main hindrance to food security, as it makes vast areas of semi-arid savannah land in Africa unsuitable for breeding domestic animals that are a source of diary and meat products [5]. In addition, AAT remains a setback in most livestock dependent economies in sub Saharan Africa, causing economic losses of agricultural productivityofabout$4.5billionperyear (AU – IBAR, 2010) and because most livestock rearing is commonly practiced by rural communities, AAT impedes rural development[6]
In livestock, AAT is caused by Trypanosoma congolense, Trypanosoma vivax, Trypanosomaevansi, Trypanosoma simiae and Trypanosomabruceibrucei [7]. Trypanosoma spp infection in cattle is primarilyspreadthroughbitesofinfected tsetse flies found in over 37 African countries in Sub Saharan Africa including Uganda known as the tsetse belt [8].Other biting flies such as Stomoxyshave also been implicated in the spread of AAT although they lack epidemiological significance[9].Anumberofhostfactors, includingphysiologicalstatusofthehost, nutritional and environmental factors, have an important effect on the pathogenicity and determine the severity of the disease [10]. Animals’ illness with T. congolense, T. vivax or T. b. brucei is characterized by fluctuating parasitaemia with periods of paroxysms and intermission. This leads to anemia, roughness of hair coat, abortion, reduced milk yield, intermittent pyrexia, depression and gradual loss of condition leading to extreme emaciation and death oftheanimal[11].
Diagnosis is an important factor in the controlling of infection. Several diagnostictechniquesfor trypanosomiasis exist, however, only few tests have been objectively analyzed and standardized, and currently fewer are mostly used in Uganda. Trypanosomiasis has traditionally been diagnosed using microscopy to directly observe the Trypanosoma parasites in blood either through a wet film system for detecting mobile trypanosomes oras thickand thin dried smears. Recently, the explosion of
Aziz et al
new techniques, because of the rapid developments in molecular biology, new approaches and technologies can be used for diagnosis [12]. The use of molecular methods such as polymerase chain reaction(PCR)hashelpedtodiagnoseand classify Trypanosoma species[13]
Thereareseveralmethodsfordetecting tr ypanosomiasis inanimals,includingparas itological,immunologicalandmolecular methods [14; 15]. This study will use traditional microscopy and molecular techniquestoquantifytheprevalenceand identifythecirculatingspecies.
Statement of the problem
In livestock dependent economies in SubSaharan Africa, Animal African Trypnosomiasis remains a threat to food security and economic livelihoods. AAT causes economic losses estimated at 3 –4.5billions USD in terms of agricultural productivity annually [1]. Lira district in northernUgandaisanactivefocusofAAT [16]
AAThasanegativeimpactonfarmersasi nfestedanimalssufferfromweightloss,r educed productivity,abortionandoftenmortality ,resultingineconomiclossesandimpedi ngsocio-economicdevelopment. Despite this, there is limited information on the prevalence and circulating trypanosomes species thus hindering efforts to manage, control eventually eradicate the AAT in Lira district and Uganda. This study therefore seeks to determine the prevalence of AAT in Lira district using microscopy and PCR technique as well as ascertain the circulating species in order to assist AAT control programs in the district.
Aim of the study
This work aim to determine the prevalence of AAT in Lira district using two selected diagnostic methods: microscopy and ITS-polymerase chain reaction
Specific objectives
1. ToassesstheprevalenceofAATof cattle in Lira district using microscopy
2. To confirm the prevalence of AAT ofcattleinLiradistrictusingPCR
3. To determine the circulating
www.idosr.org
Trypanosome spp in cattle in Lira districtusingITS-PCR
Research questions
1. What is the prevalence of AAT of cattleofcattleinLiradistrict?
2. What are the circulating Trypanosome spp in cattle in Lira district? Justification of the study
Thisstudywillgiveinformationaboutthe prevalence of Trypanosomiasis in Lira
Study Area
Aziz et al
districtwhichmayassistpolicymakersin setting up relevant anti- Trypanosomiasis interventions programs in the district. This will contribute to better animal health intervention through identification of different trypanosomes giving opportunities for advancement in Trypanosomiasis control strategy by providing basic knowledge about the species and distribution within Lira District.
METHODOLOGY
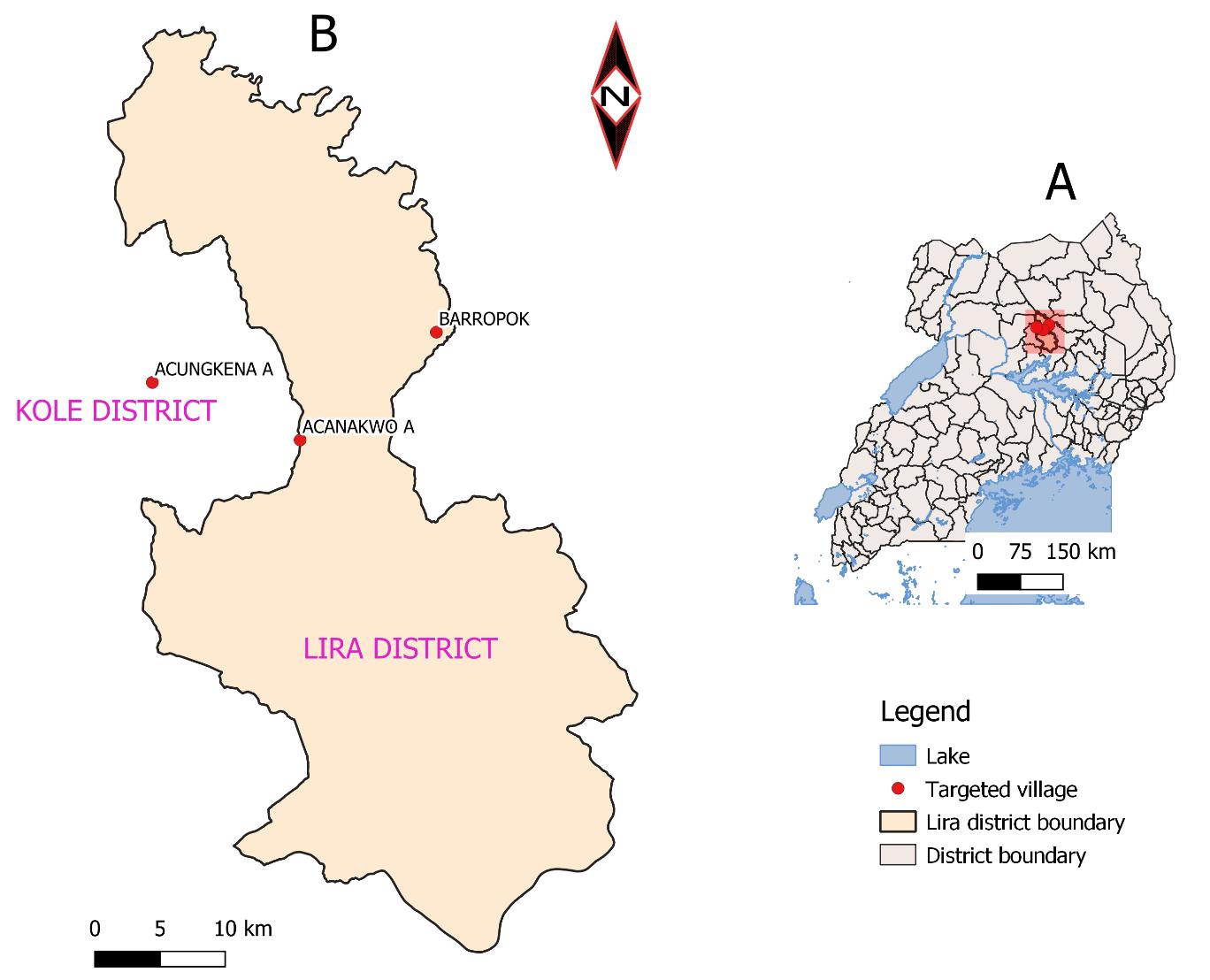
This study took place in Lira District, Uganda (Figure 1), which is located in the following coordinates: 2°14'50.0"N 32°54'00.0"E (Latitude: 02.2472; Longitude: 32.9000) in Northern Uganda [17]. Lira District is the main economic hub in northern Uganda and livestock keeping is one of the main economic activities. Furthermore, Lira District is
one of the Districts that benefitted from therestockingprogramfundedbyAfrican development bank (ADB). In 2008 the national livestock census reported the cattle population to be 15,933. This put Lira District among the cattle rearing Districts which rendered it being an AAT hotspot. The cattle were sampled from Acungkena, Barropok and Acanakwo A, sub-counties.
Sample Size Determination
This study used a survey formula previously reported by Kish [18]; n = z2p (1-p)/d2. Where: z = Z score for 95% confidence interval = 1.96, p = prevalence, d = acceptable error (5%). We used the prevalence of bovine African trypanosomes15.3% in Tororo District, south-easternUganda[19].
Study Design
This was a cross-sectional study that aimed at establishing the prevalence of Animal African Trypanosomes in Lira District using two selected diagnostic methods. A Simple random sampling technique was used to identify the study sites. Two hundred and fifty-four (254) animals were selected and assigned unique numbers for identification according to the village and farm that
they came from. This study was carried outfromMarch2020toApril2020.
Sample collection
From each selected animal, blood was collected from the jugular vein into vacutainer tubes containing EDTA and stored at 4ºC in a cooler box and transported to the District laboratory for microscopy analysis and sample processing for molecular analysis. For molecular analysis, blood from the Vacutainers tubes was applied onto Whatman FTA cards and left to dry and thenstoredinsealedplasticbagsatroom temperature. To have higher chances of detecting parasites; blood sample collectionwasdoneinthemorning[20].
Microscopy
For microscopy, 50µl of whole blood was used to make thin and thick blood films. The blood films were allowed to air dry
www.idosr.org
and then fixed in concentrated methyl alcoholforabout1-2minutesfollowedby staining with 10% Giemsa for 25-35 minutes. The blood films were then washedwithdistilledwatertoremove the excessstainandtheslideswerelefttoair dry before viewing under an oil immersionlens[21]
DNA extraction
DNA was extracted with slight modification as previously described by [5]. A 6 mm diameter disc was cut from the blood spot on the Whatman FTA card and placed into the 0.5 ml microcentrifuge tube. 200μl of sterile water was added and the tube incubated at 37°C for 30min in a heating block. The tubeswerethencentrifugedathighspeed (14000 RPM) for 1 minute and the resulting supernatant was collected and discarded. A second 100μl aliquot of sterile water was added to the Whatman FTA card discs in the original tube and incubated at 100°C for 30min in the heating block. After incubation, the sample was centrifuged at 14000 RPM for 1 minute. The supernatant was removed and stored at -20°C ready for PCR analysis.
Polymerase Chain Reaction (PCR) PCR was performed with slight modification as previously reported by
Aziz et al
[22] using a 25μl reaction volume containing 1gm of Taq (10mM Tris- HCl, 50mM KCl, 1.5mM MgCl2, 2.5 units of pureTaqDNA polymerase,200μMofeach of the four deoxynucleoside triphosphates and reaction buffer), 1μl of each primer (1μM), 2μl DNA sample and doubledistilledwatertoafinalvolumeof 25μl. Trypanosoma brucei brucei reference DNA was used as a positive control and nuclease-free PCR water was used as negative template controls. The cycling conditions that were used are; first denaturation at 98°C for 1 min, denaturation at 98°C for 5seconds, annealing at 64°C for 30 seconds, extension at 72°C for 30 seconds (25 cycles) and the final extension for 72 0 C for 10 mins [22]. The PCR conditions and cyclingconditionswerethesameforboth the 1st and 2nd PCR runs. The amplified PCR products were run on a 2% agarose gel,stainedin0.5μg/mlEthidiumbromide solution and viewed using a UV illuminator and documented. The sequences of the primers used in the amplification and expected base pairs size of each Trypanosoma species are shownin Table 1 below:
Data analysis
Ethical consideration
Ethics Committee.
www.idosr.org
for the care of animals with the help of a veterinaryassistant.
RESULTS
Prevalence of trypanosome infections in cattle
A total of 254 cattle from Acungkena, Barropok and Acanakwo A, sub-counties were screened. Of these, 107 cattle were positive for trypanosomes by hematocrit centrifugation technique (HCT), (Table 2). There was a variation in the sample size between the two sub-counties because of thecattlepopulationsperSubCounty. The prevalence of trypanosomiasis according to microscopic results was 5/254 (2.0%) as compared to 11/254 (4.3%) trypanosomiasis prevalence accordingtoPCRanalysis(Table3and5). Microscopy demonstrated that Barropok village had the highest prevalence of trypanosomiasis with 3/5 (60.0%) followedbyAcungkenavillage1/5(20.0%) andAcanakwoAvillage1/5(20.0%)(Table 3).
When the PCV results were compared to trypanosomiasis status, T. congolense distribution between normal and abnormal PCV range distribution was 6/9 (66.7%) and 3/9 (33.3%) respectively (Table 2). Infection with T. vivax and mixed infection (T. congolense/T.vivax)
Aziz et al
showed 1/1 (100.0%) trypanosomiasis status in the normal and abnormal PCV ranges respectively. Non-infected animals demonstrated 21/243 (8.6%) and 222/243 (91.4%) in the abnormal and normal PCV rangesrespectively(Table2).
Polymerase chain reaction amplification demonstrated that the prevalence of trypanosomiasis infection in the animal studied is 11/254 (4.3%). Barropok village had the highest proportion of trypanosomiasis with 6/11 (54.5%) followed by Acanakwo A village2/11 (18.2%) and Acungkena village 3/11 (27.3%) (Table 4). Trypanosoma congolense was the most dominant Trypanosomes species with a proportion of 9/11 (81.8%), followed by T.vivax 1/11 (9.1%) and mixed infection of T.congolense/T.vivax1/11 (9.1%) (Table 5 andFigure5).
Thereisstatisticalsignificantrelationship (OR=6.041; 95% CI: 1.634-22.331; p<0.05) between abnormal PCV and trypanosomiasis infection. This result also indicates that the animal with a PCV volume ≤29 is at risk of trypanosomiasis infection(Table6).
www.idosr.org
Aziz et al
Table 4: Distribution of trypanosomes species in the villages using the PCR technique
Villages T. congolense T. vivax T.congolense/T.vivax Negative Total AcanakwoA 2(3.0) 0(0.0) 0(0.0) 64(97.0) 66(26.0)
Barropok 5(4.1) 0(0.0) 1(0.8) 116(95.1) 122(48.0)
Acungkena 2(3.0) 1(1.5) 0(0.0) 63(95.5) 66(26.0) Total 9 (3.5) 1 (0.4) 1 (0.4) 243 (95.7) 254 (100.0)
Footnote:n=number,%=percentage
Table 5: Proportion of trypanosoma species
Trypanosomes species Frequency N (%) T.congolense 9(81.8) T.vivax 1(9.1) T.congolense/T.vivax 1(9.1) Total 11 (100.0)
Footnote:n=number,%=percentage
Table
Footnote: CI=confidence interval, p=probability, OR=odds ratio, p≤0.05 value is statistically significantunderlogisticregression
M N P 1 2 3 4 5 6 7 8 9 10 11 12 13 14 480bp
720bp 250bp
500bp 200bp
Figure 1: ElectrophoreticanalysisofITSPCRproducts
Representative Agarose gel results showing the different sizes of the ITS1 region for the different trypanosome species (T. brucei - 480bp, T. vivax250bp and T. congolense - 700bp) amplified using non-nested primers
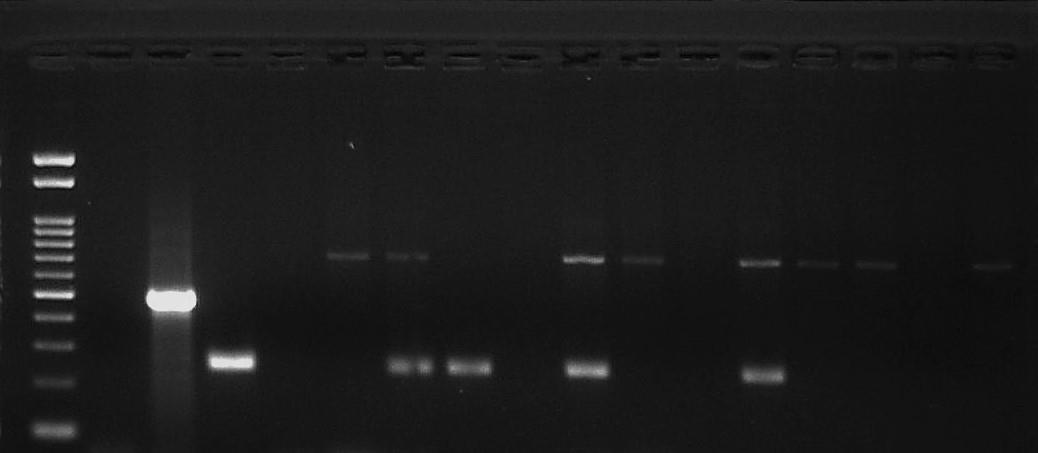
Trypanozoon that gives an expected band sizeof480bpwasnotdetected. LaneM –100bp DNA marker, Lane N – Negative control (Sterile water with no template added), Lane P – Positive control (T.b.b GVR-35),Lanes1-14 – testsamples.
700bp
DISCUSSION
Trypanosomiasis is one of the leading haemoparasitic diseases found in animals reared at home. Trypanosomiasis is transmitted by the tsetse fly species knownastheGlossinaand its widespread all over the tropical regions of Africa that exhibit vector prevalence. The main trypanosome species transmitted by the tsetse fly are Trypanosomacongolense,T. vivax and T. brucei in sheep, goats and cattleand T.simiae inpigs.Howeverlittle Knowledge about the prevalence and diversity of trypanosomes circulating in upcountry Districts in northern Uganda which lies in the tsetse fly belt [8] has been a problem making AAT management hard. Moreover, the effectiveness of AAT diagnosticmethodsusedpopularlybythe Animal health professionals in those northern Districts is not known. This study, therefore, compared the presence of animal African trypanosomes in cattle in Lira District using microscopy and molecularmethods. Theprevalenceoftrypanosomesaccording tomicroscopyresultswas5/254(2.0%)as compared to 11/254 (4.3%) trypanosomiasis prevalence according to PCR analysis. The microscopic results of this study are comparable to reports elsewhere [22; 23] and the PCR results that demonstrated a slightly higher prevalence in this study agrees with previous reports [24; 23]. The higher prevalence of trypanosomiasis shown by using the PCR method is because the moleculartechniqueismoresensitiveand specific as compared to the conventional microscopic methods [25,37,38] The substantial sensitivity of the PCR method over the conventional microscopic method in this study conforms to a previousstudy[24;25].Moststudieshave used microscopy as the reference standard. However, microscopy has relatively low sensitivity although most studies have used centrifugation to upsurgesensitivity[25,32,36,38,40]
Trypanosoma congolense was the most common Trypanosoma species with 9/11 (81.8%), followed by T.vivax 1/11 (9.1%) and mixed infection of T.congolense/T.vivax1/11 (9.1%). The high
Aziz et alproportion of T. congolense 9/11 (81.8%) demonstrated in this study as compared to T. vivax 1/11 (9.1%) is in agreement with previous reports [19] Trypanosoma congolense is known to be the leading aetiology of trypanosomiasis of the salivarian livestock group that is highly prevalent in Sub-Saharan Africa and resultsintoahugenegativeimpactonthe economy on the affected countries where it’s endemic [26,27,28,29]. This parasite causes a disease called Nagana, which means “depressed spirit” according to the ZuluofSouthAfrica.
Thisstudyalsoshowedasignificantlylow prevalence of 1/11 (9.1%) of mixed infection caused by T.congolense/T.vivax. The significantly low prevalence of 1/11 (9.1%) of mixed infection prevalence demonstrated is conformity with other previous reports [23,30,31,32] The presence of mixed infection in this study is supported by previous studies done elsewhere [26; 27,33,34,35,36]. Though the low prevalence of mixed infection demonstrated in this is not in line with otherprevious studies in otherregions of Africa where a high prevalence of trypanosomiasis due to mixed infections have been reported [24; 19,37,38]. The variations in the prevalence of mixed infections involving different species of trypanosome from one place to another are probably due to the accessibility of trypanosome species to their specific hosts. Even though little is known about the factors determining the spread and abundance of trypanosome species, the presence of a suitable mammalian host remainsthemostlikelyfactor[28,39,30]. In addition to T. congolense species with the highest proportion of 9/11 (81.8%) among AAT disease complex in the Lira District, the current investigation revealed 1/11 (9.1%) and 1/11 (9.1%) proportions of T.vivaxand T. congolense/T.vivax, respectively. This research further demonstrates that T. brucei brucei, T.vivax, and T.congolense are still present in livestock reservoirs in the broader Lango sub-region, of the Lira District [29]. The AAT disease complex in cattle is generally transmitted cyclically
www.idosr.org
by Glossina species tsetse flies. Trypanosoma vivax, on the other hand, can be spread by a wide range of haematophagous insects. A high proportion of T. vivax has been reported in south-western Uganda. Trypanosoma vivax ismoredifficulttomanagethanthe otherspecies sinceithaspreviouslybeen known to be mechanically transmitted by haematophagous insects, making it difficult to control even in regions with intensive vector control strategies [6]. A report of potential transmission of T. vivax byhematophagousinsectsinsoutheastern Uganda has been documented on the doorstepofLiraDistrict[19].To date, there has been no report of mechanical transmission of T. vivax by haematophagousinsectsinLiraDistrict. Several causes, including the gradual deterioration in state veterinary services, particularly due to adverse legislation, as well as insecurity in some parts of Uganda, may be contributing to the persistence of trypanosome infection in cattle [30]. Acaricide/insecticide and trypanocidal medicine affordability is another issue, with farmers who can afford the treatments being less likely to report cases of trypanosomiasis than those who cannot. Other likely factors include communal cattle grazing, which has been linked to a high risk of trypanosomiasis infections [17]. The continuous transmission will be achievable when cattle are grazed communally in the presence of a suitable vector because some of the cattle in the communal system will preserve the parasites. Furthermore, in addition to cattle serving as key reservoirs of trypanosomiasis, wild ungulates also play an important part in the epidemiological cycle of trypanosomiasis by serving as reservoirs. The Lango sub-region that includes Lira District still contains an
Aziz et al
abundance of free-roaming wild ungulates, which are potential trypanosomiasis reservoirs, due to its closeness to the Murchison Falls ConservationArea[29]. Polymerase chain reaction amplification demonstrated that Barropok village had thehighestprevalenceoftrypanosomiasis with 6/11 (54.5%) followed by Acanakwo A village2/11 (18.2%) and Acungkena village 3/11 (27.3%). Microscopy as well demonstrated the highest trypanosomiasis status in Barropok. This is of high concern, therefore measures should be put in place to control the tsetse flies that transmit this infection andaswelltreattheinfectedanimals. Non-infected animals demonstrated 21/243 (8.6%) and 222/243 (91.4%) in the abnormal and normal PCV ranges respectively. Most of the healthy animals had normal PCV ranges. This finding is comparable with studies done elsewhere [31]. When PCV was used as a predictor valuefortrypanosomiasisinfectionandit was subjected to bivariate analysis, it gave these logistic regression values (OR=6.041; 95% CI: 1.634-22.331; p<0.05) demonstrating a statistically significant relationship between trypanosomiasis infectionstatuswithabnormalPCVrange. The finding of this study is in agreement withotherpreviousreports[31;32].
Contribution to Scientific Knowledge
The current study has shown a trypanosomiasis prevalence of 11/254 (4.3%) in Acanakwo A, Barropok and Acungkena villages in Lira District, Uganda. This demonstrated Trypanosoma congolense as the most predominant Trypanosoma specieswithaproportionof 9/11(81.8%)inAcanakwoA,Barropokand Acungkena villages in Lira District, Uganda.
CONCLUSION
The prevalence of trypanosomiasis in Acanakwo A, Barropok and Acungkena villages in Lira District, Uganda according to PCR analysis was 11/254 (4.3%). With 9/11 (81.8%), the most common trypanosome species was Trypanosoma congolense. The highest proportion of
trypanosomiasis infection with 6/11 (54.5%) was recorded in Barropok village. Polymerasereactionamplificationwasthe mostreliablediagnosticmethodduetoits high sensitivity and specificity as compared to the conventional microscopicmethod.
www.idosr.org
Recommendations
Polymerase reaction amplification appears to have adequate accuracy to substitute the use of a microscope where facilitiesallow.
Aziz et al
Thereisurgentneedforlocalsurveillance schemesmoreespeciallyatthegrassroots in Uganda to provide data for reference guidelines development needed for controloftrypanosomiasisinUganda.
REFERENCES
1. Muhanguzi,D.,Picozzi,K.,Hattendorf, J., Thrusfield, M., Kabasa, J. D., Waiswa, C. and Welburn, S. C. (2014). The burden and spatialdistributionof bovine African trypanosomes in small holder crop- livestock production systems in Tororo District , 1–10. http://doi.org/10.1186/s13071-0140603-6
2. World Health Organization.(2014).World health statistics 2014.World Health Organization.https://apps.who.int/iri s/handle/10665/112738
3. Maigari, A. K. (2017). Soft Options for Effective Diagnosis of African Animal Trypanosomiasis:AReview, 2(2),1–9.
4. Chitanga, S., Marcotty, T., Namangala, B., Van den Bossche, P., Van Den Abbeele, J. and Delespaux, V. (2011). High prevalence of drug resistance in animal trypanosomes without a history of drug exposure.PLoS Neglected Tropical Diseases, 5(12), e1454.
5. El-Sayed, N. M., Hegde, P., Quackenbush, J., Melville, S. E. and Donelson, J. E. (2000).The African trypanosome genome. International journal for parasitology, 30(4), 329345.
6. Alingu,R.A.,Muhanguzi,D.,MacLeod, E., Waiswa, C. and Fyfe, J. (2014). Bovine trypanosome species prevalence and farmers' trypanosomiasis control methods in south-western Uganda. Journal of the South African Veterinary Association, 85(1),01-05.
7. Musinguzi, S. P., Suganuma, K., Asada, M.andLaohasinnarong,D.(2016).Full PaperParasitologyAPCR-basedsurvey of animal African trypanosomosis and selected piroplasm parasites of cattle and goats in Zambia, 4–9.journal of veterinarymedicne.160240
8. Katiti, D. (2014). Animal African trypanosomaisis and associated
cytokine profiles in naturally infected cattle in Paicho and Lakwana Subcounties, Gulu District (Doctoral dissertation,MakerereUniversity).
9. Široký, P., Kubelová, M., Modrý, D., Erhart, J., Literák, I., Špitalská, E. and Kocianová, E. (2010). Tortoise tick Hyalomma aegyptium as long term carrier of Q fever agent Coxiella burnetii evidence from experimental infection. Parasitology research, 107(6),1515-1520.
10.Uilenberg, G. and Boyt, W. P. (1998). A fieldguideforthediagnosis,treatment and prevention of African animal trypanosomosis. Food & Agriculture Org..
11.Eisler, M. C., McDermott, J. J. and Mdachi, R. E.(2000). Rapid method for the assessment of trypanocidal drug resistance in the field. Proc 9th Symp Int Soc Vet Epidemiol Econ Nairobi, Pap3539:1–3.
12.Simwango, M., Ngonyoka, A., Nnko, H. J., Salekwa, L. P., Ole-neselle, M., Kimera, S. I. and Gwakisa, P. S. (2017). Molecular prevalence of trypanosome infections in cattle and tsetse flies in theMaasaiSteppe,northernTanzania, 1–11. http://doi.org/10.1186/s13071017-2411-2
13.Alves, W. P., AbrÃ, D., Fern, L. and Facury-Filho, E. J. (2017). Comparison of three methods for diagnosis of Trypanosoma (Duttonella) vivax in cattle. Genetics and Molecular Research, 16(4).
14.Osório, A. L. A. R., Madruga, C. R., Desquesnes, M., Soares, C. O., Ribeiro, L. R. R. and Costa, S. C. G. D. (2008). Trypanosoma (Duttonella) vivax: its biology, epidemiology, pathogenesis, and introduction in the New World-a review. MemóriasdoInstitutoOswaldo Cruz, 103(1),1-13.
15.Ramírez-Iglesias,J.R.,Eleizalde,M.C., Gómez-Piñeres, E. and Mendoza, M. (2011).Trypanosoma evansi: A
www.idosr.org
comparative study of four diagnostic techniques for trypanosomosis using rabbit as an experimental model. Experimental parasitology, 128(1),91-96.
16. Nakayima,E (2016). Achieving the SDGs†:FundamentalsforUganda.
17.Von Wissmann, B., Fyfe, J.,Picozzi,K., Hamill, L., Waiswa, C. and Welburn, S.C. (2014). Quantifying the association between bovine and human trypanosomiasis in newly affected sleeping sickness areas of Uganda. PLoS Negl Trop Dis, 8(6), p.e2931.
18.Kish, L. (1965). Survey Sampling. New York: John Wiley and Sons, Inc, 275–276.
19.Muhanguzi, D, Picozzi, K, Hatendorf, J., Thrusfield, M., Welburn, S. C. and Kabasa, J. D. (2014) Improvements on Restricted Insecticide Application Protocol for Control of Human and Animal African Trypanosomiasis in Eastern Uganda. PLoS Negl Trop Dis 8(10): e3284. https://doi.org/10.1371/journal.pntd. 0003284
20.Greig, W. A., Murray, M., Murray, P. K. and McIntyre, W. I. M. (1979). Factors affecting blood sampling for anaemia and parasitaemia in bovine trypanosomiasis. British Veterinary Journal, 135(2),130-141.
21.Kirchhoff, L. V., Votava, J. R., Ochs, D. E.andMoser,D.R.(1996).Comparison of PCR and microscopic methods for detecting Trypanosoma cruzi Journal of Clinical Microbiology, 34(5), 11711175.
22.Njiru, Z. K., Constantine, C. C., Guya, S., Crowther, J., Kiragu, J. M., Thompson, R. C. A. and Dávila, A. M. R.(2005).TheuseofITS1rDNAPCRin detecting pathogenic African trypanosomes. Parasitology Research, 95(3),186-192.
23.Kivali, V., Kiyong'a, A. N., Fyfe, J., Toye, P., Fèvre, E. M. and Cook, E. A. (2020). Spatial Distribution of Trypanosomes in Cattle From Western Kenya. Frontiers in Veterinary Science, 7,554.
Aziz et al
24.Matovu, E., Mugasa, C. M., Waiswa, P., Kitibwa, A., Boobo, A. and Ndung’u, J. M.(2020).Haemoparasiticinfectionsin cattle from a trypanosoma brucei rhodesiense sleeping sickness endemic district of eastern Uganda. Tropical medicine and infectious disease,5(1),24
25.Mugasa, C. M., Adams, E. R., Boer, K. R., Dyserinck, H. C., Büscher, P., Schallig, H. D. and Leeflang, M. M. (2012). Diagnostic accuracy of molecular amplification tests for human African trypanosomiasis systematic review. PLoS neglected tropicaldiseases, 6(1),e1438.
26.Radwanska,M.,Vereecke,N.,Deleeuw, V., Pinto, J., &Magez, S. (2018). Salivarian Trypanosomosis: A Review of Parasites Involved, Their Global Distribution and Their Interaction With the Innate and Adaptive Mammalian Host Immune System. Frontiersinimmunology, 9.
27.Truc,P.,Jamonneau,V.,N'Guessan, P., N'Dri, L., Diallo, P. B. and Cuny, G. (1998). Trypanosoma brucei ssp. and T. congolense: mixed human infection in Cote d'Ivoire. Transactions of the RoyalSocietyofTropicalMedicineand Hygiene, 92(5),537-538.
28.Nimpaye, H, Njiokou, F, Njine, T, Njitchouang, G. R., Cuny, G., Herder, S., Asonganyi, T. and Simo, G. (2011) Trypanosoma vivax, T. congolense Bforest type^ and T. simiae: prevalence in domestic animals of sleeping sickness foci of Cameroon. Parasite18:171–179
29.Wangoola,R.M.,Kevin,B.,Acup,C.A., Welburn, S., Waiswa, C. and Bugeza, J. (2019). Factors associated with persistence of African animal trypanosomiasis in Langosubregion, northern Uganda. Tropical animal health and production, 51(7), 20112018.
30.Bugeza, J., Kankya, C., Muleme, J., Akandinda, A., Sserugga, J., Nantima, N. and Odoch, T. (2017). Participatory evaluationofdeliveryofanimalhealth care services by community animal health workers in Karamoja region of Uganda. PloSone, 12(6),e0179110.
www.idosr.org
31.Dagnachew, S., Terefe, G., Abebe, G., Sirak, A., Bollo, E., Barry, D. and Goddeeris, B. (2015). Comparative clinico-pathological observations in young Zebu (Bosindicus) cattle experimentally infected with Trypanosoma vivax isolates from tsetseinfestedandnon-tsetseareasof Northwest Ethiopia. BMC veterinary research, 11(1),307.
32.Yayeh, M., Dagnachew, S., Tilahun, M., Melaku, A., Mitiku, T., Yesuf, M. and Kefyalew, H. (2018). Comparative experimental studies on Trypanosoma isolates in mice and response to diminazeneaceturate and isometamidium chloride treatment. Heliyon, 4(2),e00528.
33.Hussein Osman Ahmed, Joy Muhumuza and Musiime James Nabaasa (2022). Evaluation of the factors associated with immediate adverse maternal outcomes among referred women in labor at Kampala International University Teaching Hospital. IAA Journal of BiologicalSciences8(1):228-238.
34.Hussein Osman Ahmed, Joy Muhumuza and Musiime James Nabaasa (2022). Factors associated with Immediate Adverse Maternal Outcomes among Referred Women in Labor attending Kampala International University Teaching Hospital. IAA Journal of Applied Sciences8(1):117-125.
35.Hussein Osman Ahmed, Joy Muhumuza and Musiime James Nabaasa (2022). The composite immediate adverse maternal outcomes among women in labor referred to Kampala International University Teaching Hospital IAA
Aziz et al
Journal of Scientific Research 8(1):149-156.
36.Daniel Asiimwe, Herman Lule and Izimba Daniel (2022). Epidemiology of Assault Injuries among Trauma Patients Presenting at Kampala International University Teaching Hospital and Jinja Regional Referral Hospital. INOSR Applied Sciences 8(1):111119.
37.E.O.Ikuomola,O.S Akinsonmisoye, R.O. Owolabi and M. B. Okon (2022). Assessment of Toxicity Potential of Secnidazole on Reproductive System of Male WistarRats. INOSRAppliedSciences 8(1):120-133.
38.Ugwu Okechukwu, P. C., Onwe, S. C. and Okon, M. B. (2022). The effect of Methanol Extract of Rauwolfia vomitoria on Lipid Profile of Chloroform intoxicated Wistar Albino Rats. IAA Journal of ScientificResearch,8(1),73-82.
39.E.O.Ikuomola,O.S.Akinsonmisoye, R.O. Owolabi and M. B. Okon (2022).Evaluation of the Effect of Secnidazole on Sperm Motility, Morphology, Viability and Total SpermCountof WistarRats. INOSR Experimental Sciences 8(1): 74-83, 2022.
40.E.O. Ikuomola , O.S Akinsonmisoye, R.O. Owolabi and M.B.Okon(2022).Evaluationofthe effect of secnidazole on the histology of the testes and epididymis of male Wistar rats. INOSR Experimental Sciences 8(1): 84-94,2022.
