Abstract
Climatically extreme regions such as the polar deserts of the McMurdo Dry Valleys (78° S) in Continental Antarctica are key areas for a better understanding of changes in ecosystems. Therefore, it is particularly important to analyze and communicate current patterns of biodiversity in these sensitive areas, where precipitation mostly occurs in form of snow and liquid water is rare. Humidity provided by dew, clouds, and fog are the main water sources, especially for rock-dwelling crustose lichens as one of the most common vegetation-forming organisms. We investigated the diversity and interaction specificity of myco-/photobiont associations of 232 crustose lichen specimens, collected along an elevational gradient (171–959 m a.s.l.) within the McMurdo Dry Valleys. The mycobiont species and photobiont OTUs were identified by using three markers each (nrITS, mtSSU, RPB1, and nrITS, psbJ-L, COX2). Elevation, positively associated with water availability, turned out to be the key factor explaining most of the distribution patterns of the mycobionts. Pairwise comparisons showed Lecidea cancriformis and Rhizoplaca macleanii to be significantly more common at higher elevations and Carbonea vorticosa and Lecidea polypycnidophora at lower elevations. Lichen photobionts were dominated by the globally distributed Trebouxia OTU, Tr_A02 which occurred at all habitats. Network specialization resulting from myco-/photobiont bipartite network structure varied with elevation and associated abiotic factors. Along an elevational gradient, the spatial distribution, diversity, and genetic variability of the lichen symbionts appear to be mainly influenced by improved water relations at higher altitudes.
Similar content being viewed by others
Introduction
The McMurdo Dry Valleys (MDV) in Southern Victoria Land, Continental Antarctica, are characterized by an environment that is exceptional even for Antarctica: it is extremely arid and cold, which makes it hostile for most organisms. Thus, life is rare within the valleys of this polar desert, and only few life forms can cope with these extreme conditions (e.g., Adams et al. 2006; Pointing et al. 2009). The main limiting factor for life within the MDV is water availability with fog, clouds, dew, and ephemeral melting water of snow patches having important effects on the climatic conditions (Pannewitz et al. 2005; Adams et al. 2006; Green et al. 2007; Stichbury et al. 2011). Among the most diverse macro-organisms present in the MDV are lichens. Lichens represent a classic example of symbiosis, consisting of a fungus (mycobiont) and one or more photosynthetic partners (photobiont). When completely desiccated, lichens are dormant and can survive unfavorable conditions for long periods (Kappen and Valladares 2007; Green 2008). Consequently, they are able to colonize rocks and boulders above melting streams or in the vicinity of snow patches, even in such extreme environments as the MDV (e.g., Hertel 2007; Ruprecht et al. 2010, 2012a; Schroeter et al. 2010; Green et al. 2011b; Sancho et al. 2017). The most successful species are green-algal lichens, as they do not depend on the presence of liquid water for reactivation and can be active below zero degrees, in contrast to cyanobacterial lichens that appear to be completely absent in continental Antarctica (Lange et al. 1986; Schlensog et al. 1997; Kappen 2000; Seppelt et al. 2010; Green et al. 2011a).
Several studies on mycobiont–photobiont interactions in lichens have shown that green-algal as well as cyanobacterial lichenized fungi can show considerable photobiont variability and can have more than one photobiont and even combine green alga and cyanobacteria (Nelsen and Gargas 2009; Otalora et al. 2010; Wornik and Grube 2010; Fernández-Mendoza et al. 2011; Henskens et al. 2012; Ruprecht et al. 2014; Engelen et al. 2016). Low photobiont specificity and a high ability to accept different photobionts might be a survival strategy and extend the ecological range of lichens (Wirtz et al. 2003; Blaha et al. 2006; Ruprecht et al. 2012a; Leavitt et al. 2015; Dal Grande et al. 2017). Furthermore, photobiont selection appears to be influenced by abiotic factors like climate (Beck et al. 2002; Yahr et al. 2006; Fernández-Mendoza et al. 2011; Peksa and Skaloud 2011). At the local scale (for instance along elevational gradients), this may translate into habitat-specific photobiont switches (Vargas Castillo and Beck 2012). Above all, temperature has often been identified as a key factor for photobiont selection of lichens in Antarctica (Kappen and Valladares 2007; Green 2008; Ruprecht et al. 2012a). In warmer regions, myco-/photobiont interactions show increased specificity leading to one-to-one interactions in contrast to more generalist interactions in colder environments (Singh et al. 2017). Thus, it appears that symbiotic interactions in lichens can react very sensitively to environmental change although this conclusion is based on a small database, and these responses have been investigated only in a few species (Colesie et al. 2014b; Allen and Lendemer 2016; Sancho et al. 2017). In general, there is agreement that climatic changes will influence the diversity, abundance, and growth of lichens (Sancho et al. 2017) and that lichens therefore represent excellent bioindicators for processes associated with global warming (Alatalo et al. 2015; Allen and Lendemer 2016; Bassler et al. 2016; Sancho et al. 2019).
Over the last decades, studies on elevational gradients have re-emerged because the species composition changes remarkably with elevation suggesting a species-specific adaptation to different environmental conditions (Grytnes et al. 2006). They provide steep ecological transitions (e.g., in temperature, humidity, and UV radiation) over short distances (Körner 2007; Keller et al. 2013) and several studies suggest that the structure and diversity of communities, the abundance and distribution of species, and ecosystem properties and processes can change along elevational gradients (Wolf 1993; Körner 2003; Grytnes et al. 2006; Bassler et al. 2016; Dal Grande et al. 2017; Junker and Larue-Kontic 2018). For lichens, elevational gradients are reported to show large changes in species composition (Leavitt et al. 2015; Dal Grande et al. 2017), habitat-specific photobiont switches (Vargas Castillo and Beck 2012), and/or microclimatic partitioning of ecologically differentiated fungal and algal gene pools (Nadyeina et al. 2014).
This study focuses on saxicolous crustose lichens in continental Antarctica which are associated with green micro alga as photobionts. In general, these lichens are slow growing and restricted to microhabitats on rock surfaces (Hertel 1998), but nevertheless, due to their poikilohydric nature, they are well adapted to habitats with high insolation and with rapid fluctuations in temperature and water availability (Lange 1997, 2000; Green et al. 2002; Schroeter et al. 2011). Most of the lichens analyzed here belong to the “lecideoid” lichen group (Hertel 1984) and these species are assigned to the generic name Lecidea sensu Zahlbruckner (1925) but they do not necessarily belong to the genus in its strict sense. Due to their inconspicuous growth form, distinguishable only by a few small morphological traits such as spore size and ascus type, the identification of these lichens is difficult even under best growing conditions (Ruprecht et al. 2020). Extreme climate conditions in cold deserts result in reduced development of the thallus such as chasmolithic growth, a lack of ascomata, or sparsely developed ascospores, all features that hamper identification and even the detection of the specimens in the vast landscape (Hertel 2009; Ruprecht et al. 2010). Nevertheless, these pioneers on rocks and pebbles (Hertel 1984, 1987) belong to one of the most abundant species groups in continental Antarctica (Hertel 2007; Ruprecht et al. 2010, 2012b) and are, therefore, an excellent study system to investigate changes in symbiotic associations along gradients. The present study covers lecideoid lichen species of the genera Carbonea Hertel, Lecanora Ach., Lecidella Körb., Rhizoplaca Zopf, and the genus Lecidea Ach s.str. (Hertel 1984) plus, in addition, lichen samples of the genera Austrolecia Hertel, Buellia De Not., and Huea C.W. Dodge & G.E. Baker which were included, because they often have a similar appearance.
The aim of this study was to analyze the spatial pattern and factors that might affect the distribution of both symbiotic partners within the MDV. We addressed the following objectives: (1) to confirm and extend our knowledge of the abundances of the myco- and photobiont species that have been found by previous, less extensive studies, (2) to investigate the variability of myco-/photobiont interactions, in particular, analyze the level of selectivity by using network statistics, and (3) to study for mycobionts, photobionts and lichens the relationships with abiotic and climatic factors such as elevation and water status.
Materials and methods
Study area and sampling sites
This study was conducted as part of the New Zealand Terrestrial Antarctic Biocomplexity Survey (nzTABS, https://nztabs.ictar.aq), which was initiated during the International Polar Year 2007–2008, and drew a diverse range of international expertise to profile the biology, geochemistry, geology, and climate of the MDV. The study is among the most comprehensive landscape-scale biodiversity surveys undertaken and includes nearly all trophic components found in the MDV ecosystem (Lee et al. 2019). Sampling of soils and biological communities was carried out over two successive Austral summers (2009/10, 2010/11). The geographic area within which lichen samples were collected was the southern part of the MDV (total area: 22,700 km2, ice-free area: 4500 km2; Levy 2013; Fig. 1a, b). The landscape is a mosaic of glacially formed valleys with intervening high ground, ice-covered lakes, ephemeral streams, arid rocky soils, ice-cemented soils, and surrounding glaciers along the steep scree and boulder slopes (Fig. 1b–d; Doran et al. 2002; Stichbury et al. 2011; Yung et al. 2014). There are four main valleys (Miers Valley, Garwood Valley, Hidden Valley, and Marshall Valley) and some other extensive ice-free areas (Shangri-La). The topography ranges from sea level to more than 2000 m a.s.l. with granite being the dominant rock type on the ridges and hills, while the valley floors are covered with glacial drift. The valleys have the typical glaciated form with a U-cross-section with steep sides, often with scree slopes, which reach up to around 600 m in height. To the west, the valleys are separated from the polar plateau by the Royal Society Range that has peaks over 4000 m a.s.l. in height. To the east, the valleys open out onto the Ross Ice Shelf, which represents a climatically maritime influenced location within the MDV, despite the absence of ice-free sea at any time of the year (Yung et al. 2014).
a Antarctic continent; investigated area marked with red rectangle (Natural Earth, Qgis), b MDV sampling sites defined in four areas (https://www.gpsvisualizer.com), c Garwood Valley and field camp, d Garwood Valley with incoming cloud bank, e Lecidea cancriformis, f Rhizoplaca macleanii, g Austrolecia sp. 1
Climate of the McMurdo Dry Valleys
The climate of the MDV is, for several reasons, classified as that of a polar desert. First, the mountains at the west are sufficiently high to block seaward flowing ice from the East Antarctic ice sheet from reaching the Ross Sea. In addition, the Transantarctic Mountains provide a precipitation shadow, causing an extremely low humidity and lack of snow or ice cover in the MDV (Monaghan et al. 2005). Annual precipitation is < 50 mm a−1 water equivalent, with precipitation decreasing away from the coast (Fountain et al. 2010). The major source of liquid water is the seasonal melting of perennial snowbanks and glaciers (Stichbury et al. 2011; Head and Marchant 2014) but, in most cases, this water is not available for lichens that inhabit rock surfaces above the surrounding ground level. MDV climate is best known from the northern valleys, particularly Taylor Valley, because of the McMurdo Long-Term Ecological Research programme (McMurdo LTER) that has been active since 1993 (https://www.mcmlter.org). The valley floors of the MDV show annual mean temperatures that range from − 30 to − 15 °C and typically have fewer than 50 days a−1 where average temperatures exceed 0 °C (Doran et al. 2002; Ochyra 2008; Colesie et al. 2014b). There is agreement that the air temperature lapse rate is close to 1 °C decline per 100 m elevation rise, as well as an increase with distance inland from the coast of 0.09 °C per 1 km (McKay 2015). The aspect of the valley slopes has an important impact: north facing slopes are warmer and dryer, south facing slopes are cooler and wetter (Yung et al. 2014). The wind regime is strongly topographically channeled and directed mainly up- or downvalley. During summer, easterly valley winds dominate, due to differential surface heating between the low albedo valley floors and the high albedo ice to the east (Mckendry and Lewthwaite 1990). In winter, wind direction is typically more variable. Cold air pools associated with light winds and very low minimum temperatures (− 50 °C) often occupy topographic low points of the valleys during winter (Doran et al. 2002).
Almost all climate information comes from studies on the valley floors. There are, however, conditions that tend to produce a major difference in water regime between valley floors and intervening mountain ranges. First, there is a tendency at higher elevations for greater snowfall and higher humidity, as shown by the presence of clouds at higher elevations (Fig. 1d). Second, there is the regular occurrence within the valleys of what have traditionally been regarded as katabatic winds (Mckendry and Lewthwaite 1990; Ayling and McGowan 2006) but which are now suggested to be foehn winds albeit generated in a slightly different manner to the classic northern hemisphere foehns (Speirs et al. 2010). In the Taylor Valley, for example, these winds are easily recognizable by their sudden arrival, high intensity (around 15 m s−1), rapidly rising temperature (by around 25 °C to reach about 0 °C), and rapidly falling relative air humidity to around 20% (Speirs et al. 2010). These foehn winds also occur in the southern valleys with an example from Miers Valley (Online Resource 2a) showing almost identical characteristics to those in the Taylor Valley. Foehn winds are extremely drying with air vapor pressure deficit rising about 50 times from 0.01 kPa (− 30 °C, 80% RH) to 0.49 kPa (0 °C, 20% RH). They are also topographically constrained within the valleys and can apparently reach altitudes up to almost 500 m (Speirs et al. 2010). The net result of the higher elevation cold, moister air, and the extremely drying foehn winds within the valleys is that the wetness availability gradient is strongly non-linear and, for the purposes of our analyses, we defined an elevational threshold of about 600 m a.s.l. which marks the upper limit of the steeper valley sides.
Sample sources
The present study includes 232 lichen samples (lecideoid lichen species of the genera Carbonea, Lecanora, Lecidella, Rhizoplaca, Lecidea, and additionally Austrolecia, Buellia and Huea, which have a similar appearance under these extreme climate conditions) collected in four different areas (Hidden Valley, Miers Valley, Shangri La, and Garwood Valley) at 153 different localities with a range of aspects (N-facing slopes, flat areas, and plateaus) and elevations (171–959 m a.s.l.; Fig. 1a–g, Table 1). 154 specimens were collected between the years 2009 and 2011 by Ulrike Ruprecht and Roman Türk and are deposited at the herbarium of the University of Salzburg (SZU, Online Resource 1a). An additional 78 lichen samples from the same area were obtained from the study of Pérez-Ortega et al. (2012) excluding specimens of the genera Acarospora, Caloplaca, Polysporina, Sarcogyne, and Umbilicaria (see Online Resource 1b).
Please note that for most of the data evaluations, mycobionts, and photobionts were treated separately. In some analyses (noted in text), only mycobiont species with n ≥ 10 (min10MycoSp) were used, while others included only photobiont haplotypes with n ≥ 10 (min10PhoHap).
DNA-amplification, purification, and sequencing
Total DNA was extracted from the thallus and/or apothecia by using the DNeasy Plant Mini Kit (Qiagen) following the manufacturer’s instructions. For all samples, we sequenced and amplified the internal transcribed spacer (ITS) region of the mycobionts’ and photobionts’ nuclear ribosomal DNA (nrITS). We also amplified additional markers: for the mycobionts the mitochondrial small subunit (mtSSU) and the low-copy protein coding marker RPB1 and, for the photobionts, the chloroplast-encoded intergenic spacer (psbJ-L) and part of the cytochrome oxidase subunit 2 gene (COX2). This was done using newly developed specific primers and PCR-protocols in our project framework (Ruprecht et al. 2020).
For amplifying nrITS of the mycobiont, we used the primers ITS1 (White et al. 1990), ITS1F (Gardes and Bruns 1993), ITS1L (Ruprecht et al. 2020), ITS4 (White et al. 1990), ITS4L (Ruprecht et al. 2020), and for the photobiont 18S-ITS uni-for (Ruprecht et al. 2012a), ITS1T (Kroken and Taylor 2000), ITS1aT (Ruprecht et al. 2014), ITS4T (Kroken and Taylor 2000), and ITS4bT (Ruprecht et al. 2014). For the marker mtSSU, the primers CU6 (https://nature.berkeley.edu/brunslab/tour/primers.html), mrSSU1 (Zoller et al. 1999), mtSSU for2 (Ruprecht et al. 2010), and mtSSU rev2 (Ruprecht et al. 2010) and for RPB1, fRPB1-C rev (Matheny et al. 2002), gRPB1-A for (Matheny et al. 2002), and RPB1_for_Lec (Ruprecht et al. 2020) were chosen. For the marker COX2, COXIIf2 and COXIIr (Lindgren et al. 2014) and COX sense (Ruprecht et al. 2020) and for psbJ-L, psbF (Werth and Sork 2010), psbL-sense and psbJ-antisense (Ruprecht et al. 2014) were used. All reactions were performed as described in (Ruprecht et al. 2020). Unpurified PCR products were sent to Eurofins Genomics/Germany for sequencing.
Phylogenetic analysis
The sequences of the different marker regions listed above were assembled and edited using Geneious version 6.1.8 (Kearse et al. 2012) and aligned with MAFFTv7.017 (Katoh et al. 2002) for both symbionts. For the photobiont, the classification and labeling of the different operational taxonomical units (OTUs) followed the concept of Leavitt et al. (2015), using automatic barcode gap discovery (ABGD; Puillandre et al. 2012) implemented in the reevaluated study by Ruprecht et al. (2020).
Phylogenetic relationships of the samples of the present study were calculated from the sequences of the marker nrITS, because the other markers could not be amplified in a sufficient quantity for all species, as well as intraspecific variation was in most of the cases too low, if available; therefore, they were excluded in all following analyses using sequence data.
A maximum likelihood analysis was calculated with the IQ-TREE web server (Trifinopoulos et al. 2016), using the model selection algorithm ModelFinder (Kalyaanamoorthy et al. 2017). The Bayesian information criterion (BIC) selected for the best-fit model for the mycobiont alignment TN + I + G4 and for the photobiont K2P + I. Branch supports were obtained with the implemented ultrafast bootstrap (UFBoot) (Minh et al. 2013) (number of bootstrap alignments: 1000, maximum iteration: 1000, minimum correlation coefficient: 0.99). Additionally, a SH-aLRT branch test (Guindon et al. 2010) was performed. Each branch of the resulting tree was assigned with SH-aLRT as well as UFBoot supports; the branches with SH-aLRT < 80% and/ or UFboot < 95% were collapsed by adding the command minsupnew 80/95 to the script.
Haplotype analysis
We determined the haplotypes (h) of the different mycobiont species and photobiont OTUs by using the function haplotype() of the R package pegas (Paradis 2010) (note: the function only takes into account transversions and transitions but ignores insertions and deletions). For min10MycoSp species and photobiont OTUs with h ≥ 2 and at least two haplotypes with n ≥ 3 (Lecidea cancriformis, Lecidella greenii, Rhizoplaca macleanii, and photobiont OTU Tr_A02), haplotype networks were computed, using the function haploNet() of the R package pegas (Paradis 2010). The frequencies were clustered in 10% ranges, for example, the circles of all haplotypes making up between 20 and 30% have the same size. The haplotypes were color-coded according to their spatial distribution.
Analysis of spatial distribution
To analyze how the distribution of the lichen specimens correlated with abiotic factors, the sampling sites of the different lichen species or haplotypes in the investigated areas were compared with respect to their environmental specifics. For this, we tested the variables elevation, latitude, longitude, and the BIOCLIM variables generated by Wagner et al. (2018) providing a spatial resolution of 1 km. Since only elevation with its accompanying abiotic factors such as humidity and temperature had an effect on the data. The other variables could not be suitably assessed due to the relatively small area (data not shown). To assure a minimum group size of ten sample points, the tests only included the min10MycoSp species and min10PhoHap haplotypes.
We used nonparametric multiple comparisons for relative effects (mctp-test; function mctp() of the R package nparcomp (Konietschke et al. 2015), which conducts pairwise comparisons of all possible combinations. Considering the photobionts contain four OTUs with only two of them including more than ten individuals, the elevation of the sample sites of these two most dominant photobiont OTUs (Tr_A02 and Tr_S15) was additionally compared by conducting a nonparametric t-test, using the R function npar.t.test() of the package nparcomp (Konietschke et al. 2015).
Analysis of mycobiont–photobiont associations
The associations between mycobiont and photobiont haplotypes were analyzed by computing bipartite networks, using the R function plotweb() of the package bipartite (Dormann et al. 2008). For the bipartite network including mycobiont species and photobiont haplotypes, the indices H2′ and d′ (Blüthgen et al. 2006) were calculated. Both indices are derived from Shannon entropy. H2′ characterizes the degree of complementary specialization or partitioning among the two parties of the entire bipartite network, while d′ describes the degree of complementary specialization at species or haplotype level. They both range from 0 for the most generalized to 1 for the most specialized case and were computed using the R functions H2fun() and dfun() of the package bipartite (Dormann et al. 2008).
Phylogenetic species diversity of the interaction partners was quantified by calculating a number of further metrics listed in Table 2, including the indices net relatedness index (NRI), phylogenetic species variability (PSV), and phylogenetic species richness (PSR).
Analysis of DNA polymorphism
For each identified mycobiont and photobiont species with more than one sample, we calculated the haplotype as well as the nucleotide diversity using p-distances with DnaSP v5 (Librado and Rozas 2009). Gaps and missing data were excluded. We focused on h/n (number of haplotypes, h, divided by number of samples, n, per species), hd (haplotype diversity, the probability that two randomly chosen haplotypes are different; Nei 1987), and π (nucleotide diversity, average number of nucleotide differences per site between two randomly chosen DNA sequences; Nei and Li 1979).
In order to analyze the dependence of haplotype and nucleotide diversity values on elevation, we used the defined threshold of 600 m a.s.l. The h/n, hd, d′, and PSV of those min10MycoSp species with mean values above this threshold were grouped together and then compared to those species with mean values below 600 m a.s.l., using the R function nonpartest() of the package npmv (Ellis et al. 2017), which performs nonparametric comparisons of multivariate samples. (note: π, NRI, and PSR were excluded because of high correlations (r ≥ 0.85) with h/n (π), hd (π), d′ (π), and PSV (NRI and PSR).
Results
Phylogenetic analysis
The molecular phylogenies for the mycobiont (Online Resource 2b) and the photobiont (Online Resource 2c) are based on the marker nrITS, because the additional markers (mycobiont: mtSSU, RPB1; photobiont: psbJ-L, COX2) were not available in a sufficient quantity for all species, partly because of the implementation of the dataset (solely ITS) of (Pérez-Ortega et al. 2012). Besides, the available sequences showed for mtSSU, RBP1, and COX2, in most of the cases, very low or no variation within their taxonomical units. Only the highly variable region of the IGS (psbJ-psbL) shows mere 79.3% identical sites and an elongated sequence part on the 5′ end of the IGS in contrast to northern hemispheric specimen for the OTU Tr_A02 (Online Resource 1c). This finding is supported by a cultured specimen from Antarctica (NCBI Acc.num.: MH818851; Ruprecht et al. 2020). Unfortunately, only six sequences of 202 specimens are available. However, they are an important resource for future studies, as they contain the only multilocus information for this region. Both analyses include only accessions from the study sites (Online Resource 1a, b) to present the various species and diversity levels.
Mycobiont
The final data matrix for the phylogeny comprised 232 single sequences of the marker nrITS with a length of 610 bp and included sequences of the families LECANORACEAE, TELOSCHISTACEAE, CATILLARIACEAE, CALICIACEAE, and LECIDEACEAE. The phylogenetic tree was midpoint rooted and shows a total of 25 strongly supported clades on species level, assigned to eight genera. The backbone is not supported and therefore the topology will not be discussed. The genera Huea, Austrolecia, Buellia, and Lecidea are clearly assigned to their family level and are strongly supported. The genera Carbonea, Lecidella, and Rhizoplaca assigned to the family LECANORACEAE each form highly supported clades, but do not form an independent clade. The clade of the genus Lecanora is divided in five species (L. cf. mons-nivis, L. fuscobrunnea, L. sp. 1, L. sp. 2, and L. sp. 3), Carbonea in three species (C. vorticosa, C. sp. URm1, and C. sp. 2), Austrolecia in three species (A. sp. 1, A. sp. 2, and A. sp. 4), Buellia in four species (B. frigida, B. sp. 1, B. sp. 2, and B. sp. 3), and Lecidea in seven species (L. andersonii, L. cancriformis, L. lapicida, L. polypycnidophora, L. sp. 5, L. sp. 6, and L. UCR1). The samples allocated to the genera Lecidella, Rhizoplaca, and Huea were, in each genus, monospecific (L. greenii, Rhizoplaca macleanii, and Huea sp. 1). The taxonomical assignment of the obtained sequences was especially based on the concept of (Pérez-Ortega et al. 2012) which was extended accordingly, as well as on the following literature: Ruprecht et al. (2010, 2012b, 2020).
Photobiont
The final data matrix for the phylogeny comprised 222 single sequences of the marker nrITS with a length of 578 bp. The phylogenetic tree was midpoint rooted and shows four strongly supported clades, all of them belonging to the genus Trebouxia. They were assigned to OTU level using automatic barcode gap discovery (ABGD; Puillandre et al. 2012) according to Leavitt et al. (2015) and were included in the reevaluated dataset of Ruprecht et al. (2020). The OTUs were assigned to Tr_A02, Tr_S02, Tr_S15, Tr_S18. The OTU Tr_A02 was by far the most common, being present in 202 (91%) of our 222 samples. Photobiont sequences taken from Pérez-Ortega et al. (2012) which were labeled only with numbers were included in our system of assigning the haplotypes to the appropriate OTUs (Ruprecht et al. 2020) and therefore renamed.
Haplotype analysis
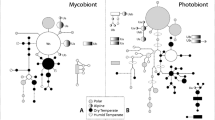
The following analyses were based on the nrITS sequences of myco- and photobionts. The number of haplotypes differed significantly between myco- and photobionts. We identified 48 different mycobiont but only 17 different photobiont haplotypes. The most frequent mycobiont haplotype was Lecidella greenii_h01 with 28 samples, the most frequent photobiont haplotype was Tr_A02_h01 with 87 samples. Additionally, some of the mycobiont species appear to be more diverse (number of different haplotypes) than others. Figures 2 and 3 show bar charts that give the number of samples per mycobiont species/photobiont OTU per area and per haplotype.
Number of samples per species/OTU and area (cf. Fig. 1b). a mycobiont species (total sample size: n = 232), b photobiont OTUs (total sample size: n = 222)
Three different mycobiont species (L. cancriformis, L. greenii, and R. macleanii) and the most common photobiont OTU (Tr_A02) met the required criteria defined above for the construction of haplotype networks (h ≥ 2 and at least two haplotypes with n ≥ 3). In Fig. 4, the respective haplotype networks show the spatial location within the four areas. As shown in Fig. 2 for mycobiont species/photobiont OTUs, the distribution again turned out to be rather uniform, with most of the haplotypes found in all of the four areas.
Haplotype networks of those mycobiont species/photobiont OTUs with h ≥ 2 and at least two haplotypes with n ≥ 3: a Lecidea cancriformis, b Lecidella greenii, c Rhizoplaca macleanii, and the photobiont OTU Tr_A02, d showing the spatial distribution within Area 1 to Area 4 (cf. Figs. 1b, 2). Roman numerals at the center of the pie charts refer to the haplotype IDs, italic numbers next to the pie charts to the total number of samples per haplotype. The circle sizes reflect relative frequency within the species/OTU; frequencies were clustered in ten, so that, for example, the circles of all haplotypes making up between 20 and 30% have the same size
Analysis of spatial distribution
For 12 of the 28 pairwise comparisons for the mycobionts species (min10MycoSp) and photobiont haplotypes (min10PhoHap), the mctp tests (pairwise comparisons of all possible combinations) for elevation showed significant differences, which are also visually recognizable when comparing the maps of the sample locations where the sample locations for each species are shown separately with varying colors indicating their elevations (min10MycoSp and min10PhoHap summarized in the respective OTUs; Online Resource 2d). The pairs with significant differences as well as the associated p-values are given in Online Resource 1d.
For the photobionts, the mctp test showed significant differences for only for two of the six pairwise comparisons (Tr_A02_h01 and Tr_A02_h03, Tr_A02_h01, and Tr_S15_h01 and, in each case, Tr_A02_h01 has lower elevation values; Online Resource 1d). The nonparametric t-test for comparing the elevation of the sample locations on OTU level resulted in a significant difference between the two groups for Tr_A02 and Tr_S15 (p = 0.005) with sampling sites of the OTU Tr_S15 being higher.
Figure 5 shows the elevational distribution of the different mycobiont species and photobiont OTUs at their sample sites for min10MycoSp species and min10PhoHap haplotypes. The mycobiont species L. cancriformis and R. macleanii had a significant tendency to higher elevations, while Carbonea vorticosa, Lecidea polypycnidophora, and L. greenii had a significant tendency to lower elevations. One haplotype of the photobiont, OTU Tr_S15, was restricted to higher elevations. The remaining mycobiont species and photobiont haplotypes have an intermediate distribution.
Analysis of mycobiont–photobiont associations
The bipartite network was calculated for all associations between the mycobiont species (min10MycoSp; lower level) and the respective photobiont haplotypes (higher level; Fig. 6). The H2′ (overall level of complementary specialization of all interacting species) of this network shows a low value of 0.226, which indicates a network with mostly generalized interactions (as opposed to specialized). This was mainly caused by the dominant occurrence of the three most abundant haplotypes (h01–h03) of the most common photobiont OTU Tr_A02 (91% of 222 accessions). All mycobiont species were additionally associated to a variety of other and less abundant haplotypes (h04–h13) of this OTU Tr_A02. Furthermore, some of the mycobionts were associated with accessions of the distantly related OTUs Tr_S02, Tr_S15, and Tr_S18. The network matrix shows the number of associations between the mycobiont species and photobiont haplotypes (Online Resource 1e). The individual d′ values (complementary specialization at species or haplotype level) are ranging from 0 to 1 and are presented in Table 3.
Bipartite network including the min10MycoSp species as well as the haplotypes of their associated photobionts with an H2′ = 0.226. Rectangles represent species, and the width is proportional to the number of samples. Associated species are linked by lines, whose width is proportional to the number of associations
Species richness of mycobiont vs. photobiont
The number of different symbiotic partners at haplotype level (SR) as a function of the number of mycobiont haplotypes (h in Table 3) is illustrated in Fig. 7 for the min10MycoSp species. We found that highly variable mycobionts are tendentially, albeit not significantly, associated with a higher number of photobiont haplotypes (Pearson correlation: t6 = 2.41, p = 0.053, r = 0.70).
Analysis of DNA polymorphism and nonparametric comparisons of multivariate samples
Analyses of DNA polymorphism and nonparametric comparisons of multivariate samples were achieved for min10MycoSp and min10PhoHap including the parameters h/n (number of haplotypes, h, divided by number of samples, n, per species), hd (haplotype diversity), and π (nucleotide diversity), d′ (specialization index), NRI (net relatedness index), PSV (phylogenetic species variability), SR (species richness), and PSR (phylogenetic species richness; Table 3).
The lowest values of hd and π were developed by four mycobiont species of min10MycoSp (C. vorticosa, L. polypycnidophora, L. greenii, and Huea sp. 1) which occur at the lowest and intermediate elevations. In contrast, species that were found only at the higher elevations (R. macleanii, L. cancriformis) showed the opposite behavior and had the highest values for hd and π. These results were supported by nonparametric tests of the different haplotype and nucleotide diversity values which showed a significant difference between min10MycoSp samples below the threshold of 600 m a.s.l. (C. vorticosa, Huea sp. 1, L. polypycnidophora, and L. greenii) and those above (Austrolecia sp. 1, Buellia frigida, L. cancriformis, and R. macleanii) in terms of h/n, hd, d′, and PSV (ANOVA type test p-value: 0.009). The scatterplots of the indices as a function of elevation as well as the Pearson correlation coefficients are presented in Online Resource 2e.
Discussion
The McMurdo Dry Valleys at 78° S in Victoria Land, Antarctica, are known as the largest continuous ice-free landscape on the continent and are mostly colonized by lithic and soil-related microbial communities (Pérez-Ortega et al. 2012; Bottos et al. 2014; Colesie et al. 2014a; De Los Rios et al. 2014; Yung et al. 2014; Lee et al. 2019). The only comprehensive evaluation to date for saxicolous lichens was that of Pérez-Ortega et al. (2012) which showed a high number of mycobiont species (26) and, in contrast, a low number of photobiont species (four). Here, we use a new and much larger dataset focused specifically on the lecideoid lichen group but also included some other species with similar morphologies in the same and adjacent areas and the respective data of Pérez-Ortega et al. (2012; Fig. 1b). A total of 25 mycobiont species were identified with a composition that was mostly similar to the previous study but including a few additional specimens (Lecanora fuscobrunnea, unknown Lecidea and Buellia species). They were all associated with the same set of common photobionts (four species), dominated (91%) by the more recently reclassified OTU Trebouxia A02 (Leavitt et al. 2015), equivalent to the species T. sp. URa2 (Ruprecht et al. 2012a). The evaluation of Pérez-Ortega et al. (2012) included not only the same set of lecideoid and morphologically similar lichen genera (Lecidea; Carbonea, Lecanora, Lecidella, Rhizoplaca; Austrolecia; Buellia; Huea) as investigated in this study, but also five additional lichen genera (Acarospora; Caloplaca; Polysporina; Sarcogyne; Umbilicaria).
In our study, we found that the different mycobiont species and photobiont OTUs within the MDV appear to be relatively evenly distributed across all four primary sample sites (Fig. 2), which is in basic agreement to the previous study. However, our results show distinct patterns for distribution, genetic diversity, and specificity. These results contrast with Pérez-Ortega et al. (2012) where the distribution of mycobionts and photobionts was independent of elevation and other abiotic factors. A clear trend has now emerged showing that the distribution of species/OTUs is significantly related to elevation, using 600 m a.s.l. as a defined threshold dividing higher and lower sites. The mycobiont species C. vorticosa, L. polypycnidophora, and L. greenii were found almost exclusively below and L. cancriformis and R. macleanii above the threshold (Fig. 5), which was supported by mctp-tests for pairwise comparisons (Online Resource 1d).
In contrast, the dominant photobiont OTU Tr_A02 is distributed everywhere while the remaining and distantly related OTUs (Tr_S02, Tr_ S15 and Tr_ S18) are mostly restricted to the higher elevated habitats (cold and humid; Fig. 5, Online Resource 2c). This result is in agreement with Dal Grande et al. (2018), who reported clear elevational preferences for some Trebouxia taxa at the OTU level at a mountain range in central Spain covering an elevational gradient of 1400 m. He suggested that elevation plays a prominent role in shaping the community structure of these algae. The distribution of Tr_S15 (equivalent to the species T. URa1), only occurring in the cold and humid areas was surprising because the first specimen of this species described was found at the climatically most extreme and dry habitats in the Darwin area at 80° S (Ruprecht et al. 2012a). However, lichen photobionts seem to have clear ecological preferences and niche differentiations. This was also shown for various Asterochloris lineages (TREBOUXIOPHYCEAE) which have diverging preferences in terms of rain and sun exposure (Peksa and Skaloud 2011) while Nadyeina et al. (2014) reported elevational partitioning in the distribution of different gene pools of the photobiont of the lungwort lichen Symbiochloris reticulate (TREBOUXIOPHYCEAE; Škaloud et al. 2016). In general, several studies suggest a strong genetic association of lichen-associated algae with climatic factors and substrate (e.g., Yahr et al. 2006; Fernández-Mendoza et al. 2011; Vargas Castillo and Beck 2012), and this has been interpreted as evidence for ecological specialization (Ruprecht et al. 2012a; Muggia et al. 2014).
The mycobiont species min10MycoSp not only show clear spatial differentiation with respect to elevation for species and OTUs but also for variables expressing the genetic diversity and specialization towards both symbiotic partners. A higher elevation correlates with a higher number of haplotypes (hd) and an increased nucleotide diversity (π) which leads to a greater intraspecific diversity within the mycobionts (Table 3). These differences are also partially reflected by a higher d′, PSV, PSR, and a low NRI which show a low relatedness to the co-occurring photobiont, associated with the rarely occurring and highly differentiated other OTUs Tr_S02, Tr_S15, and Tr_S18. Consequently, mycobionts with a high genetic diversity have a higher number of interacting partners. These findings are partially supported by the study of Singh et al. (2017), who reported climate as a selective pressure in terms of increased specificity of myco-/photobiont interactions.
Our study has also shown that there is a trend that highly variable mycobionts are associated with a larger number of photobiont haplotypes (Fig. 7). If we focus on the species which are significantly distributed either below or above the threshold of 600 m a.s.l., three main scenarios emerged: (1) mycobionts with low genetic diversity (C. vorticosa, L. polypycnidophora and L. greenii) are associated with one photobiont OTU Tr_A02 and were found in only the lower area; (2) a mycobiont with a high genetic diversity (R. macleanii) is still solely associated to one photobiont OTU (Tr_A02) and is only located at the high elevated areas; and (3) the mycobiont with the highest genetic diversity (L. cancriformis) is associated with the highest number of photobiont OTUs, in the high elevation sites. These findings are in agreement with the known distribution of L. greenii (solely associated with Tr_A02), which, so far, have only been reported for sites in the more northern parts of the Ross Sea region and have never been found at the most extreme southern environments like the Darwin area (Ruprecht et al. 2012a, b). In contrast, L. cancriformis is one of the most widespread lichens, being distributed all over Continental Antarctica and is associated with all known photobiont species (Castello 2003; Ruprecht et al. 2010, 2012a).
The above results suggest that the mycobionts are dependent on the availability of climatically adapted photobionts. However, the mycobionts seem to have also their unique climate specific preferences because they do not always make use of the whole climate niche of the associated photobionts. This seems to be the case for, e.g., L. greenii which is associated with the widely distributed photobiont (Tr_A02), but only occurring in the milder parts of the Ross Sea region. These findings are only partially in line with previous studies on macro lichens (e.g., Romeike et al. 2002; Wirtz et al. 2003) that suggest that in extreme environments like the Antarctic continent there might be a selection pressure against photobiont specificity so that more versatile mycobionts are favored. Flexibility concerning the partner choice has been considered as an adaptive strategy to survive harsher environmental conditions (Leavitt et al. 2013; Engelen et al. 2016; Singh et al. 2017; Werth and Sork 2010).
To explain the myco- and photobiont distribution, we need to understand what abiotic factors control terrestrial life in these polar ecosystems. For the MDV, Lee et al. (2019) state clearly that one of the most important factors governing the distribution of taxa in the MDV is temperature, which is accepted to be inversely correlated to elevation (McKay 2015). However, much less is known about the conditions for wetness and humidity. The available wetness index for the MDV quantifies the expected wetness of a unit within the watershed by calculating the amount of possible water flowing into that unit from estimated snow fall (Stichbury et al. 2011). For rock associated lichens this is not relevant, because they are not connected to this source of water. They are, therefore, dependent on moisture provided by the very low precipitation, infrequent melting snow (Head and Marchant 2014), and humidity provided by dew, incoming fog, and clouds from the sea. Additionally, it is now clear that the occasional foehn wind events cause severe drying within the valleys at altitudes up to 500 m. At higher elevations there is cold and moister air and this establishes a strong moisture availability gradient with elevation (Speirs et al. 2010; Fig. 1d). Our results suggest that our defined elevation threshold of about 600 m a.s.l. is a reasonable level which marks the shift from lower, dryer to higher, more humid conditions. Habitat aspect is also known to be important. Yung et al. (2014) described large differences with respect to just aspect for their chasmoendolithic microbial communities at Miers Valley. Similar results were also reported for the more maritime site, Botany Bay (Seppelt et al. 2010). We did not find any impact of other topographical features such as distance to coast, slope, aspect, and substrate. The collection sites were mainly N-facing or on plateaus, our transects were narrow and consistently only five to ten km inland plus the underlying rock in the whole area is granite and the investigated lichens are restricted to siliceous rock (Ruprecht et al. 2010, 2012b). However, our sampling was equally distributed below and above the threshold of 600 m a.s.l., so the differences found for species distribution, genetic diversity, and specificity appear to be due to the changing climate conditions, particularly moisture, along the elevational gradient.
References
Adams BJ et al (2006) Diversity and distribution of Victoria Land biota. Soil Biol Biochem 38:3003–3018. https://doi.org/10.1016/j.soilbio.2006.04.030
Alatalo JM, Jagerbrand AK, Molau U (2015) Testing reliability of short-term responses to predict longer-term responses of bryophytes and lichens to environmental change. Ecol Indic 58:77–85. https://doi.org/10.1016/j.ecolind.2015.05.050
Allen JL, Lendemer JC (2016) Climate change impacts on endemic, high-elevation lichens in a biodiversity hotspot. Biodivers Conserv 25:555–568. https://doi.org/10.1007/s10531-016-1071-4
Ayling BF, McGowan HA (2006) Niveo-eolian sediment deposits in coastal South Victoria Land, Antarctica: indicators of regional variability in weather and climate. Arct Antarct Alp Res 38:313–324. https://doi.org/10.1657/1523-0430(2006)38[313:Nsdics]2.0.Co;2
Bassler C et al (2016) Contrasting patterns of lichen functional diversity and species richness across an elevation gradient. Ecography 39:689–698. https://doi.org/10.5194/bg-13-3607-2016
Beck A, Kasalicky T, Rambold G (2002) Myco-photobiontal selection in a Mediterranean cryptogam community with Fulgensia fulgida. New Phytol 153:317–326. https://doi.org/10.1046/j.0028-646X.2001.00315.x
Blaha J, Baloch E, Grube M (2006) High photobiont diversity associated with the euryoecious lichen-forming ascomycete Lecanora rupicola (Lecanoraceae, Ascomycota). Biol J Linn Soc 88:283–293. https://doi.org/10.1111/j.1095-8312.2006.00640.x
Blüthgen N, Menzel F, Blüthgen N (2006) Measuring specialization in species interaction networks. BMC Ecol 6:9. https://doi.org/10.1186/1472-6785-6-9
Bottos EM, Woo AC, Zawar-Reza P, Pointing SB, Cary SC (2014) Airborne bacterial populations above desert soils of the McMurdo Dry Valleys, Antarctica. Microb Ecol 67:120–128. https://doi.org/10.1007/s00248-013-0296-y
Castello M (2003) Lichens of Terra Nova Bay area, northern Victoria Land (Continental Antarctica). Stud Geobot 22:3–54
Colesie C, Gommeaux M, Green TGA, Büdel B (2014a) Biological soil crusts in continental Antarctica: Garwood Valley, southern Victoria Land, and Diamond Hill, Darwin Mountains region. Antarct Sci 26:115–123. https://doi.org/10.1017/S0954102013000291
Colesie C, Green TGA, Haferkamp I, Büdel B (2014b) Habitat stress initiates changes in composition, CO2 gas exchange and C-allocation as life traits in biological soil crusts. ISME J 8:2104–2115. https://doi.org/10.1038/ismej.2014.47
Dal Grande F, Sharma R, Meiser A, Rolshausen G, Büdel B, Mishra B, Thines M, Otte J, Pfenninger M, Schmitt I (2017) Adaptive differentiation coincides with local bioclimatic conditions along an elevational cline in populations of a lichen-forming fungus. BMC Evol Biol. https://doi.org/10.1186/s12862-017-0929-8
Dal Grande F, Rolshausen G, Divakar PK, Crespo A, Otte J, Schleuning M, Schmitt I (2018) Environment and host identity structure communities of green algal symbionts in lichens. New Phytol 217:277–289. https://doi.org/10.1111/nph.14770
De Los RA, Wierzchos J, Ascaso C (2014) The lithic microbial ecosystems of Antarctica’s McMurdo Dry Valleys. Antarct Sci 26:459–477. https://doi.org/10.1017/S0954102014000194
Doran PT, McKay CP, Clow GD, Dana GL, Fountain AG, Nylen T, Lyons WB (2002) Valley floor climate observations from the McMurdo dry valleys, Antarctica, 1986–2000. J Geophys Res Atmos. https://doi.org/10.1029/2001jd002045
Dormann CF, Gruber B, Fruend J (2008) Introducing the bipartite package: analysing ecological networks. R News 8(2):8–11
Ellis AR, Burchett WW, Harrar SW, Bathke AC (2017) Nonparametric inference for multivariate data: the R package npmv. J Stat Softw 76:1–18. https://doi.org/10.18637/jss.v076.i04
Engelen A, Convey P, Popa O, Ott S (2016) Lichen photobiont diversity and selectivity at the southern limit of the maritime Antarctic region (Coal Nunatak, Alexander Island). Polar Biol 39:2403–2410. https://doi.org/10.1007/s00300-016-1915-0
Fernández-Mendoza F, Domaschke S, Garcia MA, Jordan P, Martin MP, Printzen C (2011) Population structure of mycobionts and photobionts of the widespread lichen Cetraria aculeata. Mol Ecol 20:1208–1232. https://doi.org/10.1111/j.1365-294X.2010.04993.x
Fountain AG, Nylen TH, Monaghan A, Basagic HJ, Bromwich D (2010) Snow in the McMurdo Dry Valleys, Antarctica. Int J Climatol 30:633–642. https://doi.org/10.1002/joc.1933
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118. https://doi.org/10.1111/j.1365-294X.1993.tb00005.x
Green TGA (2008) Lichens in Arctic, Antarctic and alpine ecosystems. In: Beck A, Lange OL (eds) Ökologische Rolle der Flechten. Rundgespräche der Kommission für Ökologie, Vol 36. Verlag Dr. Friedrich Pfeil, Munich, Germany, pp 45–66
Green TGA, Schlensog M, Sancho LG, Winkler BJ, Broom FD, Schroeter B (2002) The photobiont determines the pattern of photosynthetic activity within a single lichen thallus containing cyanobacterial and green algal sectors (photosymbiodeme). Oecologia 130:191–198. https://doi.org/10.1007/s004420100800
Green TGA, Schroeter B, Sancho L (2007) Plant life in Antarctica. In: Pugnaire FI, Valladares F (eds) Functional plant ecology, 2nd edn. CRC Press, Boca Raton, pp 389–433
Green TGA, Sancho LG, Pintado A, Schroeter B (2011a) Functional and spatial pressures on terrestrial vegetation in Antarctica forced by global warming. Polar Biol 34:1643–1656. https://doi.org/10.1007/s00300-011-1058-2
Green TGA, Sancho LG, Türk R, Seppelt RD, Hogg ID (2011b) High diversity of lichens at 84 degrees S, Queen Maud Mountains, suggests preglacial survival of species in the Ross Sea region, Antarctica. Polar Biol 34:1211–1220. https://doi.org/10.1007/s00300-011-0982-5
Grytnes JA, Heegaard E, Ihlen PG (2006) Species richness of vascular plants, bryophytes, and lichens along an altitudinal gradient in western Norway. Acta Oecol 29:241–246. https://doi.org/10.1016/j.actao.2005.10.007
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. https://doi.org/10.1093/sysbio/syq010
Head JW, Marchant DR (2014) The climate history of early Mars: insights from the Antarctic McMurdo Dry Valleys hydrologic system. Antarct Sci 26:774–800. https://doi.org/10.1017/S0954102014000686
Helmus MR, Bland TJ, Williams CK, Ives AR (2007) Phylogenetic measures of biodiversity. Am Nat 169:3. https://doi.org/10.1086/511334
Henskens FL, Green TGA, Wilkins A (2012) Cyanolichens can have both cyanobacteria and green algae in a common layer as major contributors to photosynthesis. Ann Bot Lond 110:555–563. https://doi.org/10.1093/Aob/Mcs108
Hertel H (1984) Über saxicole, lecideoide Flechten der Subantarktis. Beih Nova Hedwigia 79:399–499
Hertel H (1987) Progress and problems in taxonomy of Antarctic saxicolous lecideoid lichens. Bibl Lichenol 25:219–242
Hertel H (1998) Flechten im Hochgebirge. In: Jung WW (ed) Naturerlebnis Alpen (Jubiläumsschrift zum 50-jährigen Bestehen der naturkundlichen Abteilung der Sektion München im Deutschen Alpenverein E.V. Verlag Dr. F. Pfeil, München, pp 33–48
Hertel H (2007) Notes on and records of Southern Hemisphere lecideoid lichens. Bibl Lichenol 95:267–296
Hertel H (2009) A new key to cryptothalline species of the genus Lecidea (Lecanorales). Bibl Lichenol 99:185–204
Junker RR, Larue-Kontic AAC (2018) Elevation predicts the functional composition of alpine plant communities based on vegetative traits, but not based on floral traits. Alp Bot 128:13–22. https://doi.org/10.1007/s00035-017-0198-6
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. https://doi.org/10.1038/nmeth.4285
Kappen L (2000) Some aspects of the great success of lichens in Antarctica. Antarct Sci 12:314–324. https://doi.org/10.1017/S0954102000000377
Kappen L, Valladares F (2007) Opportunistic growth and desiccation tolerance: the ecological success of poikilohydrous autotrophs. In: Pugnaire FI, Valladares F (eds) Handbook of functional plant ecology. Marcel Dekker, Inc., New York, Basel, pp 10–76
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. https://doi.org/10.1093/nar/gkf436
Kearse M et al (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Keller I, Alexander JM, Holderegger R, Edwards PJ (2013) Widespread phenotypic and genetic divergence along altitudinal gradients in animals. J Evol Biol 26:2527–2543. https://doi.org/10.1111/jeb.12255
Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO (2010) Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26(11):1463–1464. https://doi.org/10.1093/bioinformatics/btq166
Konietschke F, Placzek M, Schaarschmidt F, Hothorn LA (2015) nparcomp: an R software package for nonparametric multiple comparisons and simultaneous confidence intervals. J Stat Softw. https://doi.org/10.15488/630
Körner C (2003) Alpine plant life: functional plant ecology of high mountain ecosystems, 2nd edn. Springer, Berlin, pp 1–39
Körner C (2007) The use of “altitude” in ecological research. Trends Ecol Evol 22:569–574. https://doi.org/10.1016/j.tree.2007.09.006
Kroken S, Taylor JW (2000) Phylogenetic species, reproductive mode, and specificity of the green alga Trebouxia forming lichens with the fungal genus Letharia. Bryologist 103:645–660
Lange OL (1997) Flechten in Wüsten, Halbwüsten und Steppen. In: Schöller H (ed) Geschichte, Biologie, Systematik, Ökologie, Naturschutz und kulturelle Bedeutung, vol Kleine Senckenberg-Reihe 27. Senckenbergische Naturforschende Gesellschaft, Frankfurt am Main, pp 129–138
Lange OL (2000) Die Lebensbedingungen von Bodenkrusten-Organismen: Tagesverlauf der Photosynthese einheimischer Erdflechten. Hoppea, Denkschr Regensb Bot Ges 61:423–443
Lange OL, Kilian E, Ziegler H (1986) Water-vapor uptake and photosynthesis of Lichens: performance differences in species with green and blue-green-algae as phycobionts. Oecologia 71:104–110
Leavitt SD, Nelsen MP, Lumbsch HT, Johnson LA, St Clair LL (2013) Symbiont flexibility in subalpine rock shield lichen communities in the Southwestern USA. Bryologist 116:149–161. https://doi.org/10.1639/0007-2745-116.2.149
Leavitt SD, Kraichak E, Nelsen MP, Altermann S, Divakar PK, Alors D, Esslinger TL, Crespo A, Lumbsch HT (2015) Fungal specificity and selectivity for algae play a major role in determining lichen partnerships across diverse ecogeographic regions in the lichen-forming family Parmeliaceae (Ascomycota). Mol Ecol 24:3779–3797. https://doi.org/10.1111/mec.13271
Lee CK et al (2019) Biotic interactions are an unexpected yet critical control on the complexity of an abiotically driven polar ecosystem. Commun Biol. https://doi.org/10.1038/s42003-018-0274-5
Levy J (2013) How big are the McMurdo Dry Valleys? Estimating ice-free area using Landsat image data. Antarct Sci 25:119–120. https://doi.org/10.1017/S0954102012000727
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. https://doi.org/10.1093/bioinformatics/btp187
Lindgren H, Velmala S, Hognabba F, Goward T, Holien H, Myllys L (2014) High fungal selectivity for algal symbionts in the genus Bryoria. Lichenologist 46:681–695. https://doi.org/10.1017/S0024282914000279
Matheny PB, Liu YJJ, Ammirati JF, Hall BD (2002) Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). Am J Bot 89:688–698. https://doi.org/10.3732/ajb.89.4.688
McKay CP (2015) Testing the Doran summer climate rules in Upper Wright Valley, Antarctica. Antarct Sci 27:411–415. https://doi.org/10.1017/S095410201500005x
Mckendry IG, Lewthwaite EWD (1990) The vertical structure of summertime local winds in the Wright Valley, Antarctica. Bound Layer Meteorol 51:321–342. https://doi.org/10.1007/BF00119672
Minh BQ, Nguyen MAT, von Haeseler A (2013) Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30:1188–1195. https://doi.org/10.1093/molbev/mst024
Monaghan AJ, Bromwich DH, Powers JG, Manning KW (2005) The climate of the McMurdo, Antarctica, region as represented by one year of forecasts from the Antarctic Mesoscale Prediction System. J Clim 18:1174–1189. https://doi.org/10.1175/JCLI3336.1
Muggia L, Pérez-Ortega S, Kopun T, Zellnig G, Grube M (2014) Photobiont selectivity leads to ecological tolerance and evolutionary divergence in a polymorphic complex of lichenized fungi. Ann Bot Lond 114:463–475. https://doi.org/10.1093/aob/mcu146
Nadyeina O, Dymytrova L, Naumovych A, Postoyalkin S, Werth S, Cheenacharoen S, Scheidegger C (2014) Microclimatic differentiation of gene pools in the Lobaria pulmonaria symbiosis in a primeval forest landscape. Mol Ecol 23:5164–5178. https://doi.org/10.1111/mec.12928
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nei M, Li WH (1979) Mathematical-model for studying genetic-variation in terms of restriction endonucleases. PNAS 76:5269–5273. https://doi.org/10.1073/pnas.76.10.5269
Nelsen MP, Gargas A (2009) Symbiont flexibility in Thamnolia vermicularis (Pertusariales: Icmadophilaceae). Bryologist 112:404–417
Ochyra R (2008) The illustrated moss flora of Antarctica. Cambridge University Press, Cambridge, pp 1–685
Otalora MAG, Martinez I, O’Brien H, Molina MC, Aragon G, Lutzoni F (2010) Multiple origins of high reciprocal symbiotic specificity at an intercontinental spatial scale among gelatinous lichens (Collemataceae, Lecanoromycetes). Mol Phylogenet Evol 56:1089–1095. https://doi.org/10.1016/j.ympev.2010.05.013
Pannewitz S, Green TGA, Maysek K, Schlensog M, Seppelt R, Sancho LG, Türk R, Schroeter B (2005) Photosynthetic responses of three common mosses from continental Antarctica. Antarct Sci 17:341–352. https://doi.org/10.1017/S0954102005002774
Paradis E (2010) pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26:419–420. https://doi.org/10.1093/bioinformatics/btp696
Peksa O, Skaloud P (2011) Do photobionts influence the ecology of lichens? A case study of environmental preferences in symbiotic green alga Asterochloris (Trebouxiophyceae). Mol Ecol 20:3936–3948. https://doi.org/10.1111/j.1365-294X.2011.05168.x
Pérez-Ortega S, Ortiz-Alvarez R, Green TGA, de Los RA (2012) Lichen myco- and photobiont diversity and their relationships at the edge of life (McMurdo Dry Valleys, Antarctica). FEMS Microbiol Ecol 82:429–448. https://doi.org/10.1111/j.1574-6941.2012.01422.x
Pointing SB, Chan YK, Lacap DC, Lau MCY, Jurgens JA, Farrell RL (2009) Highly specialized microbial diversity in hyper-arid polar desert. PNAS 106:19964–19969. https://doi.org/10.1073/pnas.0908274106
Puillandre N, Lambert A, Brouillet S, Achaz G (2012) ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol Ecol 21:1864–1877. https://doi.org/10.1111/j.1365-294X.2011.05239.x
Romeike J, Friedl T, Helms G, Ott S (2002) Genetic diversity of algal and fungal partners in four species of Umbilicaria (Lichenized ascomycetes) along a transect of the Antarctic Peninsula. Mol Biol Evol 19:1209–1217. https://doi.org/10.1093/oxfordjournals.molbev.a004181
Ruprecht U, Lumbsch HT, Brunauer G, Green TGA, Türk R (2010) Diversity of Lecidea (Lecideaceae, Ascomycota) species revealed by molecular data and morphological characters. Antarct Sci 22:727–741. https://doi.org/10.1017/S0954102010000477
Ruprecht U, Brunauer G, Printzen C (2012a) Genetic diversity of photobionts in Antarctic lecideoid lichens from an ecological viewpoint. Lichenologist 44:661–678. https://doi.org/10.1017/S0024282912000291
Ruprecht U, Lumbsch HT, Brunauer G, Green TGA, Türk R (2012b) Insights into the diversity of Lecanoraceae (Lecanorales, Ascomycota) in continental Antarctica (Ross Sea region). Nova Hedwig 94:287–306. https://doi.org/10.1127/0029-5035/2012/0017
Ruprecht U, Brunauer G, Türk R (2014) High photobiont diversity in the common European soil crust lichen Psora decipiens. Biodivers Conserv 23:1771–1785. https://doi.org/10.1007/s10531-014-0662-1
Ruprecht U, Fernández-Mendoza F, Türk R, Fryday AM (2020) High levels of endemism and local differentiation in the fungal and algal symbionts of saxicolous lecideoid lichens along a latitudinal gradient in southern South America. Lichenologist 52:287–303. https://doi.org/10.1017/S0024282920000225
Sancho LG, Pintado A, Navarro F, Ramos M, De Pablo MA, Blanquer JM, Raggio J, Valladares F, Green TGA (2017) Recent warming and cooling in the Antarctic Peninsula region has rapid and large effects on lichen vegetation. Sci Rep 7:5689. https://doi.org/10.1038/s41598-017-05989-4
Sancho LG, Pintado A, Green TGA (2019) Antarctic studies show lichens to be excellent biomonitors of climate change. Diversity 11:42. https://doi.org/10.3390/d11030042
Schlensog M, Schroeter B, Sancho LG, Pintado A, Kappen L (1997) Effect of strong irradiance on photosynthetic performance of the melt-water dependent cyanobacterial lichen Leptogium puberulum (Collemataceae) Hue from the maritime Antarctic. In: Kappen L (ed) Bibliotheca Lichenologica: new species and novel aspects in ecology and physiology of lichens. Berlin, pp 235–246
Schroeter B, Green TGA, Pannewitz S, Schlensog M, Sancho LG (2010) Fourteen degrees of latitude and a continent apart: comparison of lichen activity over two years at continental and maritime Antarctic sites. Antarct Sci 22:681–690. https://doi.org/10.1017/S0954102010000647
Schroeter B, Green TGA, Pannewitz S, Schlensog M, Sancho LG (2011) Summer variability, winter dormancy: lichen activity over 3 years at Botany Bay, 77 degrees S latitude, continental Antarctica. Polar Biol 34:13–22. https://doi.org/10.1007/s00300-010-0851-7
Seppelt RD, Türk R, Green TGA, Moser G, Pannewitz S, Sancho LG, Schroeter B (2010) Lichen and moss communities of Botany Bay, Granite Harbour, Ross Sea, Antarctica. Antarct Sci 22:691–702. https://doi.org/10.1017/S0954102010000568
Singh G, Dal Grande F, Divakar PK, Otte J, Crespo A, Schmitt I (2017) Fungal–algal association patterns in lichen symbiosis linked to macroclimate. New Phytol 214:317–329. https://doi.org/10.1111/nph.14366
Škaloud P, Friedl F, Hallmann C, Beck A, Dal Grande F (2016) Taxonomic revision and species delimitation of coccoid green algae currently assigned to the genus Dictyochloropsis (Trebouxiophyceae, Chlorophyta). J Phycol 52:599–617. https://doi.org/10.1111/jpy.12422
Speirs JC, Steinhoff DF, McGowan HA, Bromwich DH, Monaghan AJ (2010) Foehn winds in the McMurdo Dry Valleys, Antarctica: the origin of extreme warming events. J Clim 23:3577–3598. https://doi.org/10.1175/2010jcli3382.1
Stichbury G, Brabyn L, Green TGA, Cary C (2011) Spatial modelling of wetness for the Antarctic Dry Valleys. Polar Res. https://doi.org/10.3402/Polar.V30i0.6330
Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ (2016) W-IQ-TREE: a fast online phylogenetic tool, or maximum likelihood analysis. Nucleic Acids Research 44:W232–W235. https://doi.org/10.1093/nar/gkw256
Vargas Castillo R, Beck A (2012) Photobiont selectivity and specificity in Caloplaca species in a fog-induces community in the Atacama Desert, northern Chile. Fungal Biol 116:665–676. https://doi.org/10.1016/j.funbio.2012.04.001
Wagner M, Trutschnig W, Bathke AC, Ruprecht U (2018) A first approach to calculate BIOCLIM variables and climate zones for Antarctica. Theor Appl Climatol 131:1397–1415. https://doi.org/10.1007/s00704-017-2053-5
Webb OC, Ackerly DD, McPeek MA, Donoghue MJ (2002) Phylogenies and community ecology. Annu Rev Ecol Syst 33:475–505. https://doi.org/10.1146/annurev.ecolsys.33.010802.150448
Werth S, Sork VL (2010) Identity and genetic structure of the photobiont of the epiphytic lichen Ramalina menziesii. https://doi.org/10.3732/ajb.0900276
White TJ, Bruns TD, Lee SB, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenies. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322
Wirtz N, Lumbsch HT, Green TGA, Türk R, Pintado A, Sancho L, Schroeter B (2003) Lichen fungi have low cyanobiont selectivity in maritime Antarctica. New Phytol 160:177–183. https://doi.org/10.1046/j.1469-8137.2003.00859.x
Wolf JHD (1993) Diversity patterns and biomass of epiphytic bryophytes and lichens along an altitudinal gradient in the Northern Andes. Ann Mo Bot Gard 80:928–960
Wornik S, Grube M (2010) Joint dispersal does not imply maintenance of partnerships in lichen symbioses. Microb Ecol 59:150–157. https://doi.org/10.1007/s00248-009-9584-y
Yahr R, Vilgalys R, DePriest PT (2006) Geographic variation in algal partners of Cladonia subtenuis (Cladoniaceae) highlights the dynamic nature of a lichen symbiosis. New Phytol 172:377–377. https://doi.org/10.1111/j.1469-8137.2006.01792.x
Yung CCM, Chan Y, Lacap DC, Pérez-Ortega S, de los Rios-Murillo A, Lee CK, Cary SC, Pointing SB (2014) Characterization of chasmoendolithic community in Miers Valley, McMurdo Dry Valleys, Antarctica. Microb Ecol 68:351–359. https://doi.org/10.1007/s00248-014-0412-7
Zahlbruckner A (1925) Catalogus Lichenum Universalis, Vol III. Verlag Gebrüder Borntraeger, Leipzig
Zoller S, Scheidegger C, Sperisen C (1999) PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen-forming ascomycetes. Lichenologist 31:511–516. https://doi.org/10.1006/lich.1999.0220
Acknowledgements
We are grateful to M. Affenzeller, A. Blaschka, G. Brunauer, and A.M. Zimmermann (Salzburg, A) for valuable advice, R. Türk (Salzburg, A) for collecting one third of the lichen samples in 2010 and providing the picture for Fig. 1d and the IDA Lab Salzburg (A; Land Salzburg, WISS 2025, 20102_F1901166-KPZ) for general support. The lichen collections were supported by grants to SCC from the New Zealand Foundation for Research, Science and Technology (UOWX0710), the New Zealand Ministry of Business, Innovation and Employment (UOWX1401), and the University of Waikato. Antarctica New Zealand is thanked for logistic support. This study was financially supported by the Austrian Science Fund (FWF): P26638_B16, Diversity, ecology, and specificity of Antarctic lichens.
Funding
Open access funding provided by Paris Lodron University of Salzburg.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We hereby declare that there are no ethical issues or conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wagner, M., Bathke, A.C., Cary, S.C. et al. Myco- and photobiont associations in crustose lichens in the McMurdo Dry Valleys (Antarctica) reveal high differentiation along an elevational gradient. Polar Biol 43, 1967–1983 (2020). https://doi.org/10.1007/s00300-020-02754-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00300-020-02754-8