Abstract
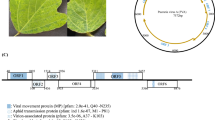
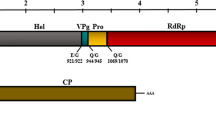
The complete genomic sequence of a waikavirus from Chinese hackberry in Zhejiang province, China, named “hackberry virus A” (HVA), was determined using high-throughput sequencing (HTS) combined with reverse transcription polymerase chain reaction (RT-PCR) and rapid amplification of cDNA ends (RACE) PCR. The bicistronic genomic RNA of HVA was found to consist of 12,691 nucleotides (nt), excluding the 3’-terminal poly(A) tail, and to encode a large polyprotein of 3783 amino acids (aa) and an additional 10.3-kDa protein. The aa sequences of the Pro-Pol and the CP regions of this virus share 39.8–44.2% and 25.5–36.4% identity, respectively, with currently known waikaviruses. These values are significantly below the current species demarcation threshold (< 75% and < 80% aa identity for the CP and Pro-Pol region, respectively) for the family Secoviridae, indicating that HVA represents a new species in the genus Waikavirus. This is the first report of a virus infecting Chinese hackberry.


Similar content being viewed by others
Data availability
All data generated during this study are included in this article.
References
Wang H, Wang S, Lan Y (2020) First report of Botryosphaeria dothidea causing leaf spot and wilt on Celtis sinensis in China. Plant Dis 17:217
Cai Y (2022) Major diseases of and control measures for Celtis sinensis. Anhui For Sci Technol 48:22–24
Weber RW (2005) Northern hackberry. Ann Allergy Asthma Immunol 94:6
Thompson JR, Dasgupta I, Fuchs M, Iwanami T, Karasev AV, Petrzik K, Sanfaçon H, Tzanetakis I, van der Vlugt R, Wetzel T, Yoshikawa N, Ictv Report Consortium (2017) ICTV virus taxonomy profile: Secoviridae. J Gen Virol 98:529–531
Seo JK, Kwak HR, Lee YJ, Kim J, Kim MK, Kim CS, Choi HS (2015) Complete genome sequence of bellflower vein chlorosis virus, a novel putative member of the genus Waikavirus. Arch Virol 160:3139–3142
Reddick BB, Habera LF, Law MD (1997) Nucleotide sequence and taxonomy of maize chlorotic dwarf virus within the family Sequiviridae. J Gen Virol 78:1165–1174
Shen P, Kaniewska M, Smith C, Beachy RN (1993) Nucleotide sequence and genomic organization of rice tungro spherical virus. Virology 193:621–630
Zhao L, Cao M, Huang Q, Wang Y, Sun J, Zhang Y, Hou C, Wu Y (2021) Occurrence and distribution of actinidia viruses in Shaanxi Province of China. Plant Dis 105:929–939
Thekke-Veetil T, Ho T, Postman JD, Tzanetakis IE (2020) Blackcurrant waikavirus A, a new member of the genus Waikavirus, and its phylogenetic and molecular relationship with other known members. Eur J Plant Pathol 157:59–64
Park D, Hahn Y (2019) A novel Waikavirus (the family Secoviridae) genome sequence identified in rapeseed (Brassica napus). Acta Virol 63:211–216
Maclot FJ, Debue V, Blouin AG, Fontdevila Pareta N, Tamisier L, Filloux D, Massart S (2021) Identification, molecular and biological characterization of two novel secovirids in wild grass species in Belgium. Virus Res 298:198397
Ito T, Sato A (2020) Three novel viruses detected from Japanese persimmon ‘Reigyoku’ associated with graft-transmissible stunt. Eur J Plant Pathol 158:163–175
Koloniuk I, Fránová J (2018) Complete nucleotide sequence and genome organization of red clover associated virus 1 (RCaV1), a putative member of the genus Waikavirus (family Secoviridae, order Picornavirales). Arch Virol 163:3447–3449
Liao R, Chen Q, Zhang S, Cao M, Zhou C (2021) Complete genome sequence of camellia virus A, a tentative new member of the genus Waikavirus. Arch Virol 166:3207–3210
Rivarez MPS, Pecman A, Bačnik K et al (2022) In-depth study of tomato and weed viromes reveals undiscovered plant virus diversity in an agroecosystem. bioRxiv 2022:498278
Tang J, Delmiglio C, Ward L, Thompson J (2022) Complete nucleotide sequence of sweetbriar rose curly-top associated virus, a tentative member of the genus Waikavirus. Arch Virol 167:651–654
Firth AE, Atkins JF (2008) Bioinformatic analysis suggests that a conserved ORF in the waikaviruses encodes an overlapping gene. Arch Virol 153:1379–1383
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A (2019) RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35:4453–4455
Acknowledgements
We thank Prof. M. J. Adams, Minehead, UK, for correcting the English of the manuscript. This work was financially supported by China Agriculture Research System of MOF and MARA (CARS-24-C-04), Ningbo Major Special Projects of the Plan “Science and Technology Innovation 2025” (2021Z106), and sponsored by K. C. Wong Magna Fund in Ningbo University.
Funding
This work were supported by Agriculture Research System of China (Grant no. CARS-24-C-04), Science and Technology Innovation 2025 Major Project of Ningbo (Grant no. 2021Z106) and K. C. Wong Magna Fund in Ningbo University.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Sead Sabanadzovic.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Weng, J., Wu, M., Ye, Z. et al. Complete nucleotide sequence of hackberry virus A, a tentative member of the genus Waikavirus. Arch Virol 168, 137 (2023). https://doi.org/10.1007/s00705-023-05764-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00705-023-05764-z