Abstract
Batoid fishes are among the most endangered marine vertebrates, yet conservation efforts have been confounded by incomplete taxonomy. Evidence suggest that the critically endangered ‘common skate’ actually represents two species: the flapper skate (Dipturus intermedius) and the blue skate (Dipturus batis). However, knowledge of the geographic range of these two nominal species is limited. Here, DNA sequencing is used to distinguish these species, allowing their spatial distributions to be clarified. These records were also used as the basis for species distribution modelling, providing the first broad scale models for each species across the Northeast Atlantic. Samples were obtained from Iceland, the UK (specifically Shetland), the North Sea and the Azores. Results suggest that D. batis was commonly distributed in the Western Approaches and Celtic Sea, extending out to Rockall and Iceland. D. intermedius generally appears to be less abundant, but was most frequent around northern Scotland and Ireland, including the northern North Sea, and was also present in Portugal. Two individuals were also identified from seamounts in remote areas of the Atlantic around the Azores, the furthest south and west the species has been found. This supports reports that the flapper skate historically had a much wider distribution (which was also highlighted in the distribution model), emphasising the large scale over which fisheries may have led to extirpations. Furthermore, these Azorean samples shared a unique control region haplotype, highlighting the importance of seamounts in preserving genetic diversity.
Similar content being viewed by others
Introduction
Elasmobranchs (sharks, skates and rays) are one of the oldest lineages of vertebrates, arising some 420 million years ago. They rapidly diverged to monopolise high trophic levels (Compagno 1990; Kriwet et al. 2008), where they may play important roles in structuring ecosystem dynamics (Heithaus et al. 2012). Typical of predatory vertebrates, elasmobranchs can be characterised by a large size, slow growth rates, late maturity, and low fecundity—exhibiting high levels of maternal investment and long gestation periods (Cortés 2000; Dulvy et al. 2014). Such life history traits, coupled with the ease of catch of larger species, make this group intrinsically at high risk of overexploitation by fisheries. In a recent global conservation assessment of chondrichthyans (sharks, skates, rays, and chimeras), approximately 25% were estimated to be threatened by extinction (Dulvy et al. 2014). In particular, batoids (skates and rays) have suffered rapid abundance declines in recent decades, with many populations suffering extirpations and significant range reductions (Brander, 1981; Casey and Meyers, 1998; Dulvy et al. 2000). Despite growing concern for their conservation status, effective management of batoids has been limited by a lack of correct species delimitation and geographic range data, often owing to the high level of morphological and ecological conservatism among extant orders (Ebert and Compagno 2007). However, mitochondrial DNA (mtDNA) has proven to be effective in reconstructing batoid phylogenies. It has also become a useful tool in establishing correct species identification, particularly when ‘cryptic’ or morphologically indistinguishable species are present within a group (Cannas et al. 2010; Griffiths et al. 2010; Iglésias et al. 2010).
The ‘common skate’, Dipturus batis (Linnaeus 1758), is one of the most vulnerable batoid species, now classified as critically endangered on the IUCN Red List (Dulvy et al. 2006). Once abundant in the Northeast Atlantic and a primary constituent of the demersal fish community, this species’ former range is thought to have stretched from Iceland and northern Norway, through to Madeira and northern Morocco, including in the Mediterranean Sea (Whitehead et al. 1984; Dolgov et al. 2005; Ellis et al. 2005). The most recent range-wide assessment by Dulvy and Reynolds (2002) indicated that the ‘common skate’ now appears to be absent from most of its former range and is locally extinct in the southern and central North Sea, western Baltic Sea and the western Mediterranean. This species has also disappeared from the Irish Sea, with just six individuals being recorded between 1988 and 1998 in the region. Notably, ‘common skate’ was the first marine fish species to have been formally described as locally extinct due to commercial fishing (Brander 1981; Dulvy and Reynolds 2002).
Recent work on the morphological, genetic, and life history characteristics of the ‘common skate’ (Iglésias et al. 2010) suggested that it actually consists of two nominal species. Concurrent work investigating the population genetic structure of ‘common skate’ around the UK drew similar conclusions, clearly distinguishing two genetically divergent and reproductively isolated groups (Griffiths et al. 2010). It was further suggested that there may be a degree of thermal and/or latitudinal separation between both groups (Griffiths et al. 2010). After a series of taxonomic revisions, the names D. intermedius (flapper skate; Parnell 1837) and D. batis (blue skate) have been adopted for these two revised species (Last et al. 2016). These results have serious implications for the conservation status of the ‘common skate’, as it is likely that the extinction risk of D. intermedius and D. batis is significantly higher than previous estimates that treated these species as a single homogenous unit. Of greatest conservation concern is perhaps D. intermedius, the larger of the two species, as size has previously been shown to be a powerful proxy for fisheries vulnerability and extinction risk in batoids (Dulvy et al. 2000; Dulvy and Reynolds 2002). However, work is still needed to clarify the distributions and abundance of these newly revised taxa. Whilst a recent study recognising the separation of these two species provided some insights into the distribution of the blue and flapper skate from around Rockall, Scotland, and as far South as the Celtic Sea (Frost et al. 2020), it did not include samples from the full extent of the range of the ‘common skate’ as reported by Dulvy and Reynolds (2002). Therefore, questions still remain as to which species, if any, is present in Iceland and the Azores (Dulvy and Reynolds 2002). Insights into the range sizes of these two species may also provide additional understanding of the scale of their declines and local extirpation.
The aim of this study was to resolve the spatial distributions of the flapper and blue skate by sampling a more comprehensive geographic range than those examined in Griffiths et al. (2010), Iglésias et al. (2010) and Frost et al. (2020). For the first time, DNA sequence data was obtained from individuals collected from Iceland, and as far south as the Azores. Mitochondrial control region (CR) sequencing was primarily used to distinguish between species. In order to facilitate broader phylogenetic comparison of these taxa, a smaller sub-set of representative samples were also analysed via the cytochrome oxidase I (COI) DNA barcode, with a focus on assessing its utility for distinguishing skate species.
Materials and methods
Control region sequence analysis
Sample collection for this study represents many years of efforts to collect blue and flapper skate tissue from across the North Atlantic. Fin clips from 44 ‘common skate’ captured during research cruises off southern Iceland in May 2011, 2012 and 2013 were collected during Norway lobster surveys (with a vessel was rigged with a conventional Nephrops trawl of 45 m headline and mesh size 80 mm), all were putatively identified as blue skate based on their morphology and were analysed with the CR sequence to confirm their identity. A further 37 skates captured from the North Sea (Shetland) and seized from a commercial boat in June 2012 were morphologically identified as the flapper skate and tissue samples were obtained for 18, which were sequenced for the CR. Samples identified simply as ‘common skate’ were also collected from long-term scientific longline surveys off the coast of the Azores (n = 2, from 2011 and 2012). Photographs (Online Resource 1) of skate in the Azores subsequently allowed the identification of these individuals as flapper skate, using morphological characteristics outlined by Iglésias et al. (2010). An additional sample captured by a commercial trawler in the eastern North Sea in 2009 was also analysed (Online Resource 2). Tissue samples were preserved in absolute ethanol prior to storage at − 20 °C. Extraction of genomic DNA was undertaken using the Promega (Madison, Wisconsin, USA) Wizard extraction kit. All individuals were sequenced for the same highly variable partial region of the mitochondrial control region, following Griffiths et al. (2010). The PCR products were sequenced by Source Bioscience (Nottingham, UK), and the results were checked by eye in BIOEDIT version 7.1.11 (Hall 1999).
To place results in a broader phylogenetic context, additional sequences from other Dipturus species were also included in the dataset (Online Resource 2). CR sequences were generated using the methods described above from four longnosed skate (Dipturus oxyrinchus; Linnaeus 1758) captured off Portugal and two barndoor skate (Dipturus laevis; Mitchill 1818) collected off the south-east Canadian coast (previously COI sequenced in Coulson et al. 2016). An additional 30 sequences were obtained from GenBank, including sequences for three additional Dipturus species (Dipturus nidarosiensis (Storm 1881), Dipturus kwangtugensis (Chu 1960), Dipturus trachyderma (Krefft and Stehmann 1975); Online Resource 3).
Cytochrome oxidase I sequence analysis
Given their rarity and the lack of knowledge surrounding the presence of the blue skate and flapper skate in the Azores (Dulvy and Reynolds 2002), the two ‘common skate’ samples from this region were sequenced for the COI gene following Ward et al. (2005). A further 76 COI sequences were also retrieved from GenBank, including an additional 12 Dipturus species (Dipturus trachyderma, Dipturus kwangtugensis, Dipturus whitleyi (Iredale 1838), Dipturus oxyrinchus, Dipturus laevis, Dipturus nidarosiensis, Dipturus cantus; last 2008, Dipturus guderi (Whitley 1940), Dipturus lemprieri (Richardson 1845), Dipturus australis (MacLeay 1884), Dipturus cerva (Whitley 1939), Dipturus confusus (Last 2008); Online Resource 4). This included two flapper skate (samples ‘12.37’ and ‘337’; GenBank accession numbers KF604223 and KF604222, respectively) and two blue skate (samples ‘12.87’ and ‘3.8’; GenBank accession numbers KF604218 and KF604220, respectively; Online Resource 2) originally from Griffiths et al. (2010). These four samples, and the two Azorean ‘common skate’ samples, represented the total number of individuals for which both the COI and CR sequences are included in the present analysis.
Due to the high level of morphological conservatism among skate species there is a possibility of misidentification of individuals in the existing datasets. Hence, when mining CR and COI sequences from GenBank a conservative approach was taken; only homologous sequences identified to the species level and from peer-reviewed publications in well-established journals were included for phylogenetic analyses. To avoid loss of phylogenetic information, sequences from GenBank that were substantial shorter than those sequenced in the current study (i.e. less than 725 and 580 base pairs for the CR and COI genes, respectively) were subsequently excluded from the analysis.
To provide an extra level of identification, COI sequences produced in the current study (from specimens AZP90 and D45 from the Azores) were also referenced against the BOLD (Barcode of Life Data System; http://www.barcodinglife.org) Identification System (BOLD-IDS), a tool that returns species -level identification from unknown 5’ COI sequences. Additionally, the grouping of these sequences into BOLD Barcode Index Numbers (BINs), which cluster barcode sequences that create operational taxonomic units closely reflective of species groupings, was examined (Ratnasingham and Hebert 2007).
Phylogenetic and phylogeographic analysis
CR and COI sequence datasets for all Dipturus species were aligned alongside those of outgroups Okamejei hollandi and Okamejei kenojei using the CLUSTALW plugin in Geneious 6.0.6 (Biomatters, Auckland, New Zealand). The most appropriate substitution model for phylogenetic analyses was determined using MEGA 7.0.26 (Kumar et al. 2016). HKY (Hasegawa et al. 1985) and K2 (Kimura 1980) were identified as the best models for the CR and COI dataset, respectively. However, the K2 model is not implemented in MrBayes or PhyML; therefore, the next most appropriate model, HKY, was selected for the COI instead. All phylogenetic reconstruction was conducted in Geneious 6.0.6. Maximum likelihood (ML) trees were constructed using the PhyML (Guindon and Gascuel 2003) plugin in. The following parameters were used: 1000 bootstrap replicates (Felsenstein 1985), an estimated gamma distribution parameter, an estimated transition/transversion ratio, proportion of invariable sites fixed at 0, 4 substitution rate categories. Bayesian phylogenetic trees were estimated using the MrBayes plugin (Huelsenbeck and Ronquist 2001); four Monte-Carlo chains were run for 1,100,000 generations, with sampling frequency set at every 200 generations. Burn-in length was set at 400,000. Sequence divergence was estimated under the gamma model, enabling the rate variation to be set at four. Consensus trees were built using the Consensus Tree Builder after removing the initial 10% burn-in; support threshold was set at 50%.
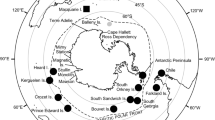
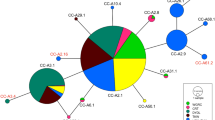
A distribution map was created using GeoMapApp© (www.geomapapp.org; CC BY Ryan et al. 2009) using all flapper and blue skate individuals analysed in the current study that had capture location information available. This totalled 65 individuals sequenced in the current study (Online Resource 2), 123 individuals sequenced in previous studies (Online Resource 3) and one individual (flapper skate) from Portugal sequenced for the COI gene, although the exact latitude and longitude for this specimen was not available (Accession number JQ774529; Online Resource 4). Therefore, the distribution map collects together the capture locations of a total of 189 individuals that have been assigned to species using mtDNA sequencing. To reconstruct genealogical relationships among haplotypes, a minimum spanning haplotype network was creating using TCS 1.2.1 (Clement et al. 1999) and tcsBU (Santos et al. 2016), with gaps selected as a fifth character state. This network was constructed using all CR sequences sequenced in the current study (Online Resource 2) and those mined from GenBank for which there was location information available (Online Resource 3), which corresponded to 188 individuals. Samples were grouped based on sea borders (World Atlas 2020): Atlantic, Celtic Sea, and North Sea. Although Rockall and Iceland are within the Atlantic, they were also grouped to reflect their separation from other Atlantic D. batis samples by significant oceanographic features, e.g. the Rockall Trough or the Maury Sea channel (Fig. 1).
a Map detailing the range of the ‘common skate’ in 2002 (adapted from Dulvy and Reynolds 2002). b Red circles represent the geographical locations of all blue skate (Dipturus batis) specimens used in phylogenetic analysis. c Green triangles represent the geographical locations of all flapper skate (Dipturus intermedius) specimens used in phylogenetic analysis. Dotted lines represent samples that were grouped together into geographic units (based on sea boarders) for the CR haplotype network. A single asterisk (*) indicates that the flapper skate specimen from Portugal did not have exact latitude and longitude information available, and so the approximate location has been indicated (as specified by Costa et al. 2012). The size of a point is proportional to the number of individuals, and the centre of each point represents the latitude and longitude for that group of specimens
Species distribution modelling
Species distribution models to predict the probability of the presence of D. batis and D. intermedius in the studied area were generated with MaxEnt v3.4.4 (Phillips et al. 2006). MaxEnt uses a maximum entropy algorithm to generate the models, considering environmental variables and species presence data (Phillips et al. 2006). All capture locations for specimens included in Fig. 1 (that were DNA sequenced to confirm their identity) were used to build the model, with the exception of one sample of D. intermedius from Portugal (accession number: JQ774529) as the exact latitude and longitude of capture was not published (Costa et al. 2012). Environmental variables including surface ice thickness mean, surface primary productivity mean, surface salinity mean, surface temperature mean, benthic maximum depth primary productivity mean, benthic maximum depth salinity mean, benthic maximum depth temperature mean, benthic minimum depth primary productivity mean, benthic minimum depth salinity mean and benthic minimum depth temperature mean were obtained using Bio-Oracle (Assis et al. 2017). The coast distance variable was generated in ArcGis 10.2 (ESRI, Redlands, USA) from the species occurrence points using the tool Euclidean distance. All variables were downloaded (or converted) to a resolution of 30 s of arc (~ 1 km2). To avoid collinearity, only variables that were not highly correlated (r < 0.8) were included.
In total, 70 different models were generated in MaxEnt for each species to select the environmental variables and MaxEnt parameters such as regularisation multiplier and features. The ENMTools software (Warren et al. 2010) was used to select the best model based on the AIC values. The final models were run with 1500 iterations using the cloclog model output. The parameters were defined as regularisation multiplier = 2 for both species, three features (linear, quadratic, and hinge) for D. batis and one feature (hinge) for D. intermedius. The reliability of the modelling result was evaluated using ten-fold cross-validations and the area under the receiver operator curve (AUC), a measure of the ability of the model to distinguish between presence locations and background/ pseudoabsences. The model AUC scores were compared with 100 null models, generated through resampling the ‘Benthic minimum depth primary productivity mean’ layer in ENMTools (Warren et al. 2010), to determine whether the models performed significantly better than random (Raes and ter Steege 2007).
Results
Phylogenetic analysis
Maximum likelihood and Bayesian trees built from COI (Fig. 2) and CR sequences (Fig. 3) showed that among the Dipturus species, most were resolved with moderate to high support (although some key nodes in each analysis were weakly supported, i.e. < 70%). In COI trees, support was low for Dipturus cerva (ML/ Bayesian: 66%/57%). Low support was also seen for Dipturus lemprieri in COI ML trees, although support was much higher in the Bayesian phylogeny (ML/ Bayesian: 55%/100%). The analyses show the ‘northern’ and the ‘southern’ clades of ‘common skate’ identified by Griffiths et al. (2010) correspond to the species flapper skate (D. intermedius) and blue skate (D. batis), respectively, and as distinguished by Last et al. (2016). Again, bootstrap support for the separation of these clades was high in both the COI and CR trees (Fig. 2, Fig. 3). Interestingly, within both the MrBayes and PhyML CR and COI trees, D. batis was not reciprocally monophyletic, grouping instead with D. laevis, the barndoor skate, a species common to the northwest Atlantic (Fig. 2). Genetic distance between these two species in trees built from both datasets was also low compared to other well-established sister species (Fig. 2, Fig. 3). Several other taxa (D. trachyderma, Dipturus kwantugensis, D. laevis, and D. nidarosiensis) were not reciprocally monophyletic in CR trees (Fig. 3). The inability to resolve some species in the phylogenetic analyses could reflect the recent divergence of species, confounded by the slow rates of sequence divergence in elasmobranchs (Dudgeon et al. 2012).
The original morphological identification of specimens on research cruises from Iceland (blue skate, D. batis), Shetland (flapper skate, D. intermedius), and the Azorean region (flapper skate, D. intermedius) was supported by the genetic analysis. Interestingly, the Azorean flapper skate revealed CR and COI haplotypes not shared by individuals in more northerly regions analysed in the current study (Fig. 2, Fig. 3). One specimen of D. intermedius from Portugal sequenced by Costa et al. (2012) also revealed a COI haplotype not identified across the other specimens included in this analysis (Fig. 2).
Comparison of the two Azorean flapper skate COI sequences with species/sequences in the BOLD-IDS revealed 100% sequence matches with five records. Four were private, but identified as D. intermedius, and the remaining record was identified as D. batis from the Northeast Atlantic (sample ID GN2181, dating from 2009, predating the recognition of D. intermedius and accounting for the use of this inconsistent scientific name). No further information regarding collection site was available for any of these records. This supports the identification of the Azorean specimens as D. intermedius and indicates that the COI haplotypes that were isolated from the Azores in the current study have been recorded previously, but their geographic origin is unknown. These sequences grouped in a BOLD BIN (AAA8675) predominately comprised of samples identified as D. intermedius or D. cf. intermedius, with a 1.68% p-distance to D. oxyrinchus as its nearest neighbour, which is consistent with the phylogenetic analysis presented here. For comparison, COI barcodes of the blue skate included in the phylogenetic analysis were also interrogated in BOLD. These were grouped into a BOLD BIN (AAC0752) that contains a mixture of records from D. batis and D. laevis. This is also consistent with the current study’s phylogenetic analysis, which failed to separate these species strongly and the very short genetic distances observed between these taxa in the trees presented here.
Phylogeographic analysis
In support of results from Griffiths et al. (2010), in the CR haplotype network, the blue skate group (haplotype diversity 0.5790) was proportionally more diverse than the flapper skate group (haplotype diversity 0.3191; Table 1; Fig. 4). The blue skate clade included 15 haplotypes shared by 129 individuals, with the most common haplotype present in all locations sampled (Iceland, Rockall, the Celtic Sea, and the Atlantic). The flapper skate clade was separated by 23 mutational steps and was represented by just three haplotypes and 59 individuals (Fig. 4), with the most common haplotype present in all sample regions, except the Azores. The distinctiveness of the Azorean flapper skate samples was supported by the haplotype network, their haplotype was three mutational steps away from the most common haplotype in this clade. For a detailed breakdown of the number of CR haplotypes per location see Online Resource 5.
Species distribution modelling
The models were generated using 49 unique location records for D. batis (shared by 129 individuals) and 27 for D. intermedius (shared by 59 individuals). Both models presented good predictive power (Table 2) and performed better than null models, AUCtrain range: 0.70–0.78. Surface primary productivity mean, benthic minimum depth primary productivity mean, and benthic minimum depth temperature mean were the main environmental variables affecting habitat suitability for the target species (Table 2). Species distribution models are shown in Fig. 5.
Discussion
A history of significant declines in abundance (Dolgov et al. 2005; Ellis et al. 2005), local extirpations (Dulvy et al. 2006) and taxonomic confusion (Griffiths et al. 2010; Iglésias et al. 2010) have left the distribution of the blue skate (D. batis) and the flapper skate (D. intermedius) uncertain, prompting the investigation of their respective distributions in the present study. The results of the current study were consistent with the conclusions of Griffiths et al. (2010) and Iglésias et al. (2010), suggesting that the known ‘common skate’ actually represents two distinct species. However, the results did not support the suggestion by Griffiths et al. (2010) that a latitudinal separation between both groups exists. Indeed, species distribution modelling suggests some overlap in habitat between these two species. For the first time, genetic data supported the occurrence of the blue skate in Iceland and the flapper skate in the Azores. The confirmation of the occurrence of flapper skate in seamounts in the Azores was a significant finding. It underlines that the distribution of D. intermedius was perhaps once much more extensive and fits with the narrative of local expiration and range-wide declines of this species. The identification of novel haplotypes from these flapper skate was also significant, highlighting the potential role of the Azores as a refugia for genetic diversity.
The integration of molecular data now permits a synthesis of spatial distribution information concerning these threatened species of skate. Since Dulvy and Reynolds’ assessment of the ‘common skate’ species complex in 2002, it is clear that division of this species has produced two groups with heavily restricted ranges (Fig. 1). The smaller species, nominally D. batis, occurs mainly in the Western Approaches and the Celtic Sea, extending out to Rockall in the Northeast Atlantic (Griffiths et al. 2010; Frost et al. 2020), with results of the current study demonstrating its occurrence around Iceland. The ‘common skate’ has long been recorded as a bycatch species in Icelandic fisheries. Historically, they were fairly common, but landings might nowadays only represent a small proportion (~ 10%) of what was landed 50 years ago (WGEF 2019). Interestingly, D. batis was not reciprocally monophyletic in COI or CR sequence phylogenies, instead samples grouped with D. laevis with shallow levels of genetic distance between the two species compared to other well-established sister species. The failure to separate these taxa has been noted in previous studies (Ball et al. 2016), and comparisons of blue skate sequences with those found in BOLD BINs showed that this species grouped into a BOLD BIN (AAC0752) containing a mixture of records from D. batis and D. laevis. In the current analysis, the COI and CR regions have not been concatenated, allowing the ability of each sequence to distinguish species to be assessed (and across the datasets key individuals and taxa lack sequences that limit the integration of these data). However, low statistical support was seen in both CR and COI trees, which has been partially overcome in previous analysis by combining these sequences (Griffiths et al. 2010). Indeed, the incorporation of other mitochondrial genes into the current dataset may also help to improve support in the phylogenetic analysis. The NADH2 gene may represent a good candidate to further resolve the mitochondrial phylogeny; due to its high level of polymorphism, it has been successfully used for species delineation in batoids (Naylor et al. 2012; Lim et al. 2015; Henderson et al. 2016). Further analysis of poorly resolved taxa, like D. batis and D. laevis, perhaps employing next-generation sequencing (NGS) and utilising greater numbers of nuclear DNA markers, may be required to resolve questions relating to the taxonomic integrity of these long-standing species.
The larger species, D. intermedius, was not found in Rockall, which is consistent with previous research (Griffiths et al. 2010; Frost et al. 2020), and inclusion of samples from Iceland here did not identify the species in this region (blue skate are more abundant in these areas). Instead, the flapper skate appears to be mainly present off Northern Ireland and Scotland (Griffiths et al. 2010; Frost et al. 2020), including the northern North Sea, with individuals less commonly found in the Celtic Sea (Frost et al. 2020). This is consistent with previous research, suggesting ‘common skate’ were regularly observed off the coast of northern and northwest Scotland, Ireland and the Celtic Sea (Dulvy and Reynolds 2002; Dulvy et al. 2006; Neat et al. 2015). Rare individuals of these species were also previously reported in the North Sea (e.g. Ellis et al. 2005; Silva et al. 2012; ICES 2012). Further north, ‘common skate’ are known to occur around Shetland (Walker and Hislop 1998; Dulvy et al. 2006), and the data presented here suggested that populations in this area were mainly comprised of flapper skate; this is consistent with previous research (Griffiths et al. 2010; Frost et al. 2020) and reports from grey literature (Shark Trust, 2009; Shark Trust, 2010). Such reports also suggest Shetland’s role in supporting significant numbers of flapper skate eggs, joining the western Scottish coast as potential nursery areas for this species (Shark Trust 2010). Identification and protection of such regions is vital in the conservation of D. intermedius. This is all the more important given the threat of scallop dredging in the region, which remains the third most important sector of the UK fishing industry (The Scottish Fishermen’s Federation 2018), and which could potentially lead to the damage or removal of skate eggs sharing the same habitat. Further investigation of the impacts of dredging on skates is needed to properly assess the conservation implications.
Historically, the range of the ‘common skate’ was described as extending much further northwards than Shetland into Scandinavia (Fig. 1). The distribution modelling for both species also suggests the suitability of habitat along the Norwegian coast. Whilst no samples of ‘common skate’ could be obtained from Norway for the present study, previous analysis of ‘common skate’ from Norwegian and Swedish museum collections, from both morphology and DNA barcoding, generally suggested the historic presence of blue skate in the region (Viinamäki 2010). More recent work also suggests misidentification may be at the root of some records of occurrence, especially in more northerly regions (Lynghammar et al. 2014).
The southern range limits of ‘common skate’ historically encompassed the island of Madeira and northern Morocco, and the Mediterranean Sea, although it is now thought to be locally extinct in the Black Sea and the Levantine Mediterranean basin (Serena 2005). It is notable that recent inspections of fish markets at ports from 2014 to 2016 also failed to demonstrate the presence of any blue or flapper skate along the Atlantic coast of Morocco (S. Hook, personal communication). However, a single specimen of ‘common skate’ from the continental shelf of Portugal (Costa et al. 2012) was identified as a flapper skate, and the two flapper skate individuals collected from seamounts in the Azores suggested this species may once have had a much wider distribution that extended into more southerly regions. Establishing the historical distributions of endangered skate is an essential part of informing conservation and IUCN Red List assessments (Dulvy et al. 2006), and these results were consistent with evidence from historical accounts of the occurrence of the ‘common skate’ (which has been hypothesised to have been confused under the name Raja macrorynchus; Rafinesque 1810; Moreau, 1881) across Europe (e.g. Clark 1926; Wells 1958; Fig. 1). These Azorean specimens could represent a relict population, insularised by increased fishing pressure and subsequent large-scale declines and extirpation of skates in waters associated with the European continental shelf (Dulvy et al. 2002). The relative inaccessibility of the Azores, lack of significant trawl fishing and the fact that this species has little commercial importance in the region, may have protected the skate from overfishing there (Abecasis et al. 2015). Furthermore, the Azores have some of the earliest designations of marine protected areas (MPAs), meaning it is likely that a population of flapper skate could have persisted. Although many of these MPAs were originally coastal and do not correspond to the deeper waters in which these specimens were captured, coastal areas have been shown to be important habitats for benthic skates (Shark Trust 2010; Abecasis et al. 2015). Given the limited haplotype diversity associated with more northern populations of flapper skate (59 specimens from the Atlantic, eastern North Sea, and Shetland shared just three CR haplotypes; Fig. 4), the identification of unique CR haplotypes from these Azorean specimens supports this conclusion and highlights the general importance of Azorean seamounts in facilitating the preservation of genetic diversity for this species (Reboleira et al. 2011). Comparisons of these Azorean COI barcodes with sequences in BOLD show other individuals with the same COI haplotype, but the geographic origins of these BOLD records remain unknown. The unique COI haplotype from the flapper skate specimen from Portugal (Costa et al. 2012) reinforces the genetic distinctiveness of southern D. intermedius populations.
The flapper skate now appears to occur mainly in northern Scotland and Ireland, with rarer individuals having been caught in the Celtic Sea (Frost et al. 2020). This is likely to be a heavily reduced range for this species, particularly as the ‘common skate’ is thought to have historically been present in much of the North Atlantic (Brander, 1981; Dulvy and Reynolds, 2002; Dolgov et al. 2005; Ellis et al. 2005). However, a note of caution should be taken when inferring the historical range of this cryptic species; due to the lack of accurate fisheries data it is impossible to know whether historical accounts of ‘common skate’ correspond to flapper skate or blue skate or both. Reported skate landings are often misidentified or are not reported on a species-specific basis. For example, doubts about the validity of historical identifications of ‘common skate’ across France and the Mediterranean region have been raised, due to potential misidentifications with the Norwegian skate (D. nidarosiensis) and the longnosed skate (D. oxyrinchus) (Dulvy et al. 2006; Iglésias et al. 2010). Nevertheless, due to its very large size (up to 2288 mm in length, Iglésias et al. 2010), low fecundity, and long period required to reach reproductive maturity (approximately 11 years; Dulvy et al. 2006), the flapper skate is probably more vulnerable to overfishing than the generally smaller blue skate (Dulvy et al. 2000; Dulvy and Reynolds 2002). This species appears to have undergone steep declines in number across its range (Iglésias et al. 2010) and eventually suffered fisheries-induced local extinctions.
The current study provides new, broader-scale distribution models for both D. intermedius and D. batis than those that have been previously published (Pinto et al. 2016; Frost et al. 2020), thereby aiming to provide a tool to inform the proper designation of conservation measures for these highly vulnerable species. Modelling shows considerable overlap of suitable habitat between flapper and blue skate at the broad Northeast Atlantic scale, despite previous work suggesting that potential differences in latitude (Griffiths et al. 2010) or temperature (Griffiths et al. 2010; Frost et al. 2020) may contribute to the separation of these species geographically. Indeed, high habitat suitability is identified for both species in northern regions of the North Sea, the coastal areas off northern Scotland, the Western Approaches, and the Celtic Sea. Suitable habitat is further identified in regions where one or both species have been reported to have largely disappeared, but historically may have been present, e.g. the Irish Sea, Norwegian Sea, northern Norway, and southern areas of the North Sea. It is also interesting that areas around Rockall, and further north around Iceland, are also highlighted as suitable habitat for both species, despite only D. batis being identified in these regions. Modelling also highlights suitable habitat for D. intermedius in southern areas, suggesting a wider distribution for this species. High habitat suitability is shown around the Portuguese coast, supporting the published barcode record of a single specimen from the continental shelf of Portugal (Costa et al. 2012) identified as D. intermedius in the current study. Further spots of suitable habitat are also identified within the Mediterranean Sea, potentially supporting the historic occurrence of this species in the region. Whilst this data provides useful insights into the distributions of the blue and flapper skate, the relatively small number and low density of capture locations used to inform habitat models in the current study could limit modelling accuracy. Further data will undoubtedly refine the accuracy of such models (Merow et al. 2013).
The present study has important implications for the management of the taxa formerly known as the ‘common skate’. A detailed geographical distribution of the two species, the blue and the flapper skate, is presented and incorporates novel data from individuals collected from as far north as Iceland and as far south as the Azores. This study also includes the first species distribution models for both species across the Northeast Atlantic. It contributes to a better knowledge of the distributions of D. batis and D. intermedius, and hence to a proper designation/implementation of conservation measures for these highly vulnerable species. Points of future research include a genetic analysis of these two species utilising a greater range of molecular markers, in order resolve patterns of fine-scale genetic structuring and investigate wider taxonomic uncertainties. Furthermore, new global conservation assessments of the flapper and blue skate are needed in the face of the new taxonomic and distributional data.
Data availability
Novel DNA sequences utilised in this study have been submitted to GenBank and all accession numbers are available in the Online Resources.
References
Abecasis RC, Afonso P, Colaço A, Longnecker N, Clifton J, Schmidt L, Santos RS (2015) Marine conservation in the Azores: evaluating marine protected area development in a remote island context. Front Mar Sci 2:104
Assis J, Tyberghein L, Bosh S, Verbruggen H, Serrão EA, De Clerck O (2017) Bio-ORACLE v2.0: extending marine data layers for bioclimatic modelling. Glob Ecol Biogeogr 27:277–284
Ball RE, Serra-Pereira B, Ellis J et al (2016) Resolving taxonomic uncertainty in vulnerable elasmobranchs: are the Madeira skate (Raja maderensis) and the thornback ray (Raja clavata) distinct species? Consv Genet 17:565–576
Brander K (1981) Disappearance of common skate Raia batis from the Irish Sea. Nature 290:48–49
Cannas R, Follesa MC, Cabiddu S, Porcu C, Salvadori S, Iglésias S, Deiana AM, Cau A (2010) Molecular and morphological evidence of the occurrence of the Norwegian skate Dipturus nidarosiensis (Storm, 1881) in the Mediterranean Sea. Mar Biol Res 6:341–350
Casey JM, Myers RA (1998) Near extinction of a large, widely distributed fish. Science 281:690–691
Clark RS (1926) Rays and skates. A revision of the European species. Fisheries Board for Scotland Scientific Investigations. H.M. Stationary Off.
Clement M, Posada D, Crandall KA (1999) TCS: a computer program to estimate gene genealogies. Mol Ecol Notes 9:1657–1659
Compagno LJV (1990) Alternative life-history styles of cartilaginous fishes in time and space. Environmental Biology of Fish I0020es. In: Bruton M.N. (Eds.) Alternative life-history styles of fishes. Developments in environmental biology of fishes (pp 33–75) Springer, Dordrecht
Cortés E (2000) Life history patterns and correlations in sharks. Rev Fish Sci 8:299–344
Costa FO, Monica L, Martins R, Costa MH, Costa ME, Carneiro M, Alves MJ, Steinke D (2012) A ranking system for reference libraries of DNA barcodes: application to marine fish species from Portugal. PloS One 7(4):e35858
Dolgov AV, Drevetnyak KV, Gusev EV (2005) The status of skate stocks in the Barents Sea. J Northwest Atl Fish Sci 35:249–260
Dudgeon CL, Blower DC, Broderick D, Giles JL, Holmes BJ, Kashiwagi T, Krück NC, Morgan JAT, Tillett BJ, Ovenden JR (2012) A review of the application of molecular genetics for fisheries management and conservation of sharks and rays. J Fish Biol 80:1789–1843
Dulvy NK, Metcalfe JD, Glanville J, Pawson MG, Reynolds JD (2000) Fishery stability, local extinctions, and shifts in community structure in skates. Conserv Biol 14:283–293
Dulvy NK, Reynolds JD (2002) Predicting extinction vulnerability in skates. Conserv Biol 16:440–450
Dulvy NK, Notarbartolo di Sciara G, Serena F, Tinti F, Ungaro N, Mancusi C, Ellis J (2006) Dipturus batis. The IUCN Red List of Threatened Species 2006: e.T39397A10198950.https://doi.org/10.2305/IUCN.UK.2006.RLTS.T39397A10198950.en. Accessed 26 January 2018
Dulvy NK, Fowler SL, Musick JA, Cavanagh RD, Kyne PM, Harrison LR (2014) Extinction risk and conservation of the world’s sharks and rays. eLife 3:e00590
Ebert DA, Compagno LJV (2007) Biodiversity and systematics of skates (Chondrichthyes: Rajiformes: Rajoidei). Environ Biol Fishes 80:111–124
Ellis JR, Dulvy NK, Jennings S, Parker-Humphreys M, Rogers SI (2005) Assessing the status of demersal elasmobranchs in UK waters: a review. J Mar Biolog Assoc UK 85:1025
Felsenstein J (1985) Confidence limits on phylogenies: an approach using bootstrap. Evolution 39:783–791
Frost M, Neat FC, Stirling D, Bendall V, Noble LR, Jones CS (2020) Distribution and thermal niche of the common skate species complex in the north-east Atlantic. Mar Ecol Ser 656:65–74
Griffiths AM, Sims DW, Cotterel SP et al (2010) Molecular markers reveal spatially segregated cryptic species in a critically endangered fish, the common skate (Dipturus batis). Proc Royal Soc B 277:1497–1503
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Hasegawa M, Kishino H, Yano T (1985) Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22:160–174
Heithaus MR, Wirsing AJ, Dill LM (2012) The ecological importance of intact top-predator populations: a synthesis of 15 years of research in a seagrass ecosystem. Mar Freshw Res 63:1039–1050
Henderson AC, Reeve AJ, Jabado RW, Naylor GJP (2016) Taxonomic assessment of sharks, rays and guitarfishes (Chondrichthyes: Elasmobranchii) from south-eastern Arabia, using the NADH dihydrogenase subunit 2 (NADH2) gene. Zool J Linn Soc 176:399–442
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
ICES (2012) Rays and skates in the North Sea. ICES Advice 2012, Book 6
Iglésias SP, Toulhoat L, Sellos DY (2010) Taxonomic confusion and market mislabelling of threatened skates: important consequences for their conservation status. Aquat Conserv 20:319–333
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kriwet J, Witzmann F, Klug S, Heidtke UH (2008) First direct evidence of a vertebrate three-level trophic chain in the fossil record. Proc Royal Soc B 275:181–186
Kumar S, Stecher G, Tamuar K (2016) MEGA7: Molecular evolutionary genetics analysis version for bigger datasets. Mol Biol Evol 33:1870–1874
Last PR, Weigmann S, Yang L (2016) Changes to the nomenclature of the skates (Chondrichthyes: Rajiformes). In book: Rays of the World - supplementary information, Chapter: 2. CSIRO Australian National Fish Collection, Australia
Lim KC, Lim P, Chong VC, Loh K (2015) Molecular and morphological analyses reveal phylogenetic relationships of stingrays focusing on the family Dasyatidae (Myliobatiformes). PLoS One 10(4):e0120518
Lynghammar A, Christiansen JS, Griffiths AM, Bakken T (2014) DNA barcoding of the norther Northeast Atlantic skates (Chondrichthyes, Rajiformes), with remarks on the widely distributed starry ray. Zool Scr 43:485–495
Merow C, Smith MJ, MJ, Silander JA, (2013) Jr practical guide to MaxEnt for modeling species’ distributions: what it does, and why inputs and settings matter. Ecography 36:1058–1069
Moreau E (1881) Histoire Naturelle des Poissons de la France, Paris, G. Masson
Naylor GJP, Caira JN, Jensen K, Rosana KAM, William W, Last P (2012) A DNA sequence–based approach to the identification of shark and ray species and its implications for global elasmobranch diversity and parasitology. Bull Am Mus Nat Hist 367:1–262
Neat F, Pinto C, Burrett I, Cowie L, Travis J, Thorburn J, Gibb F, Wright PJ (2015) Site fidelity, survival and conservation options for the threatened flapper skate (Dipturus cf. intermedia). Aquat Conserv 25:6–20
Phillips SJ, Anderson RP, Schapire RE (2006) Maximum entropy modelling of species geographic distributions. Ecol Modell 190:231–259
Pinto C, Thorburn JA, Neat F, Wright PJ, Wright S, Scott BE, Cornulier T, Travis JM (2016) Using individual tracking data to validate the predictions of species distribution models. Divers Distrib 22:682–693
Raes N, Ter Steege H (2007) A null-model for significance testing of presence-only species distribution models. Ecography 30:727–736
Ratnasingham S, Hebert PDN (2007) BOLD: the barcode of life data system (www. barcodinglife.org). Mol Ecol Notes 7:355–364
Reboleira AS, Borges PAV, Gonçalves F, Serrano ARM, Oromi P (2011) The subterranean fauna of a biodiversity hotspot region - Portugal: an overview and its conservation. Int J Speleol 40:23–37
Ryan WBF, Carbotte SM, Coplan J, O’Hara S, Melkonian A, Arko R, Weissel RA, Ferrini V, Goodwillie A, Nitsche F, Bonczkowski J, Zemsky R (2009) Global Multi-Resolution Topography (GMRT) synthesis data set. Geoch Geophy Geosyst 10:Q03014
Santos AM, Cabezas MP, Tavares AI, Xavier R, Branco M (2016) tcsBU: a tool to extend TCS network layout and visualization. Bioinformatics 32:627–628
Serena F (2005) Field identification guide to the sharks and rays of the Mediterranean and Black Sea. FAO, Rome
Silva J, Ellis T, Catchpole TL (2012) Species composition of skates (Rajidae) in commercial fisheries around the British Isles and their discarding patters. J Fish Biol 80:1678–1703
Shark Trust (2009) An illustrated compendium of sharks, skates, rays and chimaera. Chapter 1: The British Isles. Part 1: Skates and Rays
Shark Trust (2010) Shark focus. Issue 38, July 2010.
The Scottish Fishermen’s Federation. Fishing for the truth. (2018). http://www.fishingforthetruth.co.uk/environment/scallop-fishing/ Accessed 3rd April 2018
Warren DL, Glor RE, Turelli M (2010) ENMTools: a toolbox for comparative studies of environmental niche models. Ecography 33:607–611
Wise D (2010) Flapper skate conservation in the Orkney Isles. Shark Focus 39:16–17
Valsecchi E, Pasolini P, Bertozzi M, Garoia F, Ungaro N, Vacchi M, Sabelli B, Tinti F (2005) Rapid miocene-pliocene dispersal and evolution of Mediterranean rajid fauna as inferred by mitochondrial gene variation. J Evol Biol 18:436–446
Viinamäki E (2010) Systematic problems in skates (Batoidea: Rajidae) in Sweden: generic affiliation of Dipturus linteus, species determination of Rajella fyllae and the endangered Dipturus batis. Dissertation, Stokholm University
Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PD (2005) DNA barcoding Australia’s fish species. Philos Trans Royal Soc B 360:1847–1857
Ward RD, Holmes BH (2007) An analysis of nucleotide and amino acid variability in the barcode region of cytochrome c oxidase I (cox1) in fishes. Mol Ecol Notes 7:899–907
Ward RD, Holmes BH, White WT, Last PR (2008) DNA barcoding Australian chondrichthyans: results and potential uses in conservation. Mar Freshw Res 59:57–71
Wells AL (1958) The observer’s book of sea fishes. Frederick Warne and Co Ltd, London
WGEF (2019) Demersal elasmobranchs - Iceland and East Greenland. ICES Scientific Reports 1:422–434
Whitehead ML, Bauchot JC, Hureau J, Nielsen E (1984) Fishes of the North-Eastern Atlantic and Mediterranean. Volume 1. P. J. PTortonese Unesco, Paris, 510 pp.
World Atlas (2020) Europe. https://www.worldatlas.com/webimage/countrys/eu.htm. Accessed 6 February 2020
Wynen L, Larson H, Thorburn DC, Peverell S, Morgan DL, Field I, Gibb K (2009) Mitochondrial DNA supports the identification of two endangered river sharks (Glyphis glyphis and Glyphis garricki) across northern Australia. Mar Freshw Res 60:554
Acknowledgements
We thank Arve Lynghammar for his considerable expertise and insight into the status of common skate in Norway. Paul Bentzen is thanked for providing reference tissue from Barndoor skate. We appreciate Orly Razgour’s excellent guidance in approaching the spatial modelling. We also thank the reviewers for their helpful comments and feedback.
Funding
This work was supported by the University of Exeter as part of a self-funded Masters by Research (Biological Sciences) by Maisie Bache-Jeffreys.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
The majority of samples utilised in this study were collected with sampling procedures approved by the Marine Biological Association (MBA) Ethical Review Committee and were licenced by the UK Home Office under the Animals (Scientific Procedures) Act 1986. All sampling performed in Icelandic waters followed the ethics of the ministry of fisheries and innovation of Iceland and national ethical requirements. The remaining samples were collected at fish markets from fisheries products or from fisheries monitoring survey cruises in the case of the Azores samples.
Consent for publication
All authors consent for publication of this work.
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bache-Jeffreys, M., de Moraes, B.L.C., Ball, R.E. et al. Resolving the spatial distributions of Dipturus intermedius and Dipturus batis—the two taxa formerly known as the ‘common skate’. Environ Biol Fish 104, 923–936 (2021). https://doi.org/10.1007/s10641-021-01122-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10641-021-01122-7