Abstract
Nearly a century has passed since the first crosses were made between wheat (Triticum L.) and perennial Triticeae relatives with the goal of developing a perennial grain and forage crop. Numerous crosses of different species and genera have been attempted, and many have yielded fertile hybrids. Despite these successes, a definitive taxonomic treatment of stable hybrids has never been established. “Perennial wheat” is the term commonly used to refer to these hybrids when the traits of interest are the perennial growth habit and grain yield, regardless of parentage. In order to establish a consistent system in which researchers can effectively communicate and collaborate, it is important to characterize unique combinations. In this paper we briefly outline the history of perennial wheat breeding, suggest a naming convention based on the International Code for Nomenclature and describe one combination within the new nothogenus ×Tritipyrum. The development of perennial grains has the potential to allow for new agricultural systems that take advantage of the persistent nature of the crop. The taxonomic definition of this new crop type will help focus research and breeding efforts as well as organize the literature and facilitate collaboration.
Similar content being viewed by others
Introduction
Efforts to develop a perennial grain and forage crop by crossing wheat (Triticum L.) with perennial relatives began in the early part of the 20th century (White 1940). Researchers in multiple countries established the interfertility of wheat with various species of wheatgrass (mostly classified then in the genus Agropyron Gaertn.) and undertook breeding projects to combine the agronomics of the wheat crop with the persistence these wild relatives (Peto 1936; Smith 1942). The motivation was to capture some of the ecological benefits of perennial crops for grain production, which is dominated by annuals, though most of the current work is focused on introgression of resistance to biotic and abiotic stress (Franke et al. 1992; Cox et al. 2002; Hayes et al. 2012). The majority of the species in the tribe Triticeae Dumort. are perennial in habit, with annual growth the exception rather than the rule.
Some of the earliest crosses were carried out by Tsitsin in the former USSR. His initial experimentation led to the initiation of wide hybridization programs in other countries (Tsitsin and Lubimova 1959). He went as far as naming some of these hybrids, but did so without regard for the what was, at that time, the International Code for Botanical Nomenclature (ICBN), now the International Code for Nomenclature of algae, fungi and plants (ICN) and placed them as a new species of Triticum (Brickell et al. 2009; McNeill et al. 2012). Many researchers have investigated the wide hybridization of wheat and have contributed much to our understanding of the genetics and forms of resulting hybrids, but taxonomic treatment has been sporadic and inconsistent (Sax 1935; Johnson 1938; White 1940; Sando 1960; Suneson et al. 1963; Larson et al. 2012). Taxonomic treatment and appropriate nomenclature will aid in defining this new crop as it is developed.
Taxonomy
Wheat and related species have a basic chromosome number of 7 (Aase 1935). Wheat is able to hybridize with some perennial relatives and produce occasional offspring (Sharma and Gill 1983). Not all of these forms are known to exist in the wild, but they can be maintained under cultivation. The chromosomes of wheat and related grass species are considered homoeologous, meaning they share a common ancestry but are different in structure and DNA sequences. In hybrids with perennial relatives, wheat chromosomes generally do not pair and recombine with their homoeologues due to presence of the Ph system of wheat, which regulates homologous pairing (Sears 1976; Luo et al. 1996). This homoeology means that the fertile, stable offspring of the intergeneric hybrids are generally amphiploids resulting from fertilization via unreduced gametes. They are, therefore, a combination of distinct genomes from their parents, though always with the potential for translocations and other meiotic abnormalities that can result in homoeologous exchanges (Cai et al. 2001). The relative genomic contributions of each parent, and the overall genomic composition, define each hybrid genetically.
Species and generic boundaries are defined by interfertility, morphology, and functional grouping. The designator “perennial wheat” represents a functional grouping of potentially disparate organisms connected only by a shared breeding objective. Interfertility is the more relevant means of categorization for breeding as it is based on karyotypic and sequence compatibility, which translates into the capacity for genetic recombination and thus ultimately dictates the evolution of the crop.
Taxonomy is an attempt to discern the most parsimonious phylogeny for species, which is then reflected in nomenclature. There are many different ways to view living organisms in relation to one another, and each has some advantage and disadvantage. Morphological characterization, functional grouping, ability to interbreed and genomic constitution are all equally valid and correspondingly limited (Barkworth 1992; Spooner et al. 2003). Genera within the Triticeae have been subjected to repeated reorganization based on new genomic information and there remain conflicting views as to their best classification (Feuillet and Muehlbauer 2009). This is particularly true for the wild relatives of wheat, which have undergone reclassification and reinterpretation as new cytological and genomic information has been generated (Wang 1992; Liu and Wang 1993; Zhang et al. 1996; Kishii et al. 2005; Arterburn et al. 2011). As pointed out by Kellogg most recently, despite new genomic and cytological evidence, the importance of Triticum, Secale and Hordeum has meant that “maintaining nomenclatural stability for the major crops forces inherent ambiguity in the classification of the rest of the tribe (2015).”
Natural hybridization events occasionally blur the distinctions noted between different types of organisms. The chance cross of T. turgidum L. with Aegilops tauschii Coss. resulted in a distinct, stable amphiploid, T. aestivum (Goncharov 2011). Distinctions are particularly apparent in the ongoing conversation surrounding human generated, directed hybrids (Soltis and Soltis 2009). Discussion of the proper taxonomic treatment of Triticum and Secale L. hybrids has shown that opinions about the ontology of a species are varied, and at times, divisive (Gupta and Baum 1986). The Triticeae are especially difficult because, as Bernhardt points out, they “show a low barrier against hybridization and other introgressive events (2015)”, which, combined with genomic complexity, can lead to a wide range of combinations. The most important considerations for breeding, as well as nomenclature, are specificity and consistency.
A potential explanation for the reluctance to establish a clear taxonomy for perennial wheat is that focus has often been on genetic studies of hybrid materials at early generation that are still highly unstable in both phenotype and karyotype. The complexities of the genetic interactions in such lines have yet to yield repeatable patterns and the difficulties of managing the linkage drag associated with the wild parentage, while maintaining the perennial habit across generations, complicates breeding programs (Banks et al. 1993). A considerable body of work performed by many major contributors has characterized combinations of various genera, species and cultivars of wheat and wheatgrass, although most investigators were not specifically attempting to breed for stable hybrids and much knowledge and germplasm has been lost over time (Armstrong 1936; Peto 1936; White 1940; Love and Suneson 1945; Dvořák 1976; Gupta and Fedak 1986; Mujeeb-Kazi et al. 1987; Mujeeb-Kazi and Hettel 1995; Zhang et al. 1996; Fedak and Han 2005; Jauhar and Peterson 2013).
The historic fluidity of generic and morphological designation within the Triticeae makes this clarification especially important, and difficult. There have been significant reorganizations based on genomic data, and a unified concept of this tribe has not yet solidified (Dewey 1983; Löve 1984; Barkworth and Dewey 1985; Barkworth 1992; Wang 1992; Hsiao et al. 1995; Li et al. 2007; Barkworth et al. 2009). This reorganization has an impact on nothogenera, hybrids of two or more established genera, which can lead to confusion, as in the case of ×Agrotriticum Cif. et Giacom.
The nothogenus ×Agrotriticum has been used inconsistently for the stable amphiploids of Triticum and Agropyron Gaertn. senso lato, and was formed when the genus Agropyron encompassed a wide range of now separate groups (Aase 1935; Barkworth 1992; Wang 2011). It was formally designated by Ciferri and Giacomini in (1950) where they also assigned names at the species level for unique combinations, but without typification (1950). In the literature ×Agrotriticum at the genus level is confusing because the reorganization of the Triticeae has moved all but a few species from Agropyron, making it difficult to determine the genera and species of the parents based on current treatment.
Perennial wheat has been the catch-all common term used to refer to wide crosses made between different species of Triticum and related wheatgrass species, many in the Thinopyrum Á. Löve genus, towards the goal of a perennial grain crop (Suneson et al. 1963; Scheinost et al. 2001). Establishing accurate binomials for specific combinations of genera and species maintains an aspect of the pedigree with the crop and buffers that information against future reorganization of the parent species. If the parental genera or species are changed in the future, names can be updated and valuable information about the lines, and the interrelationship between species, will be retained. It also recognizes the objective of developing a new crop type that may be made up of different species, similar to Triticum.
The most relevant and direct example of a similar situation is that of Triticale (properly ×Triticosecale Wittmack ex A. Camus but frequently ×Triticale Müntzing), the result of intergenric hybridization between Secale and Triticum species (Baum and Gupta 1990). Baum (1971) advocated for different species combinations of rye (Secale) and wheat (Triticum) to be recognized as nothospecies within the nothogenus to clarify their parentage. In a review of the nomenclature of Triticale, Stace (1987) found six published names at the generic level, only two of which were valid, and 33 specific names, only two being valid. The system we propose would help avoid this type of situation.
Nomenclature
We propose that as breeders work to develop perennial wheat as a crop, they contribute also to taxonomy and nomenclature. Each unique combination should be defined, as put forth by the ICN, and given both proper nothogeneric and nothospecific rank. Each combination of genera should have one legitimate nothogenus and each combination of species from those genera one legitimate nothospecific name. If the complexity of combinations developed through breeding exhausts the provisions of the ICN the cooperation of breeders and taxonomists will be needed.
Recommendation for emendation of the ICN
Plant breeding is the art and science of creating new genetic combinations of plants. Taxononomists work to understand origins and categorize the diversity of plant species. These two actions inform each other as breeders use the result of taxonomic study to push genetic boundaries for crop improvement. It may be that an emendation to the ICN, as suggested by both Stace (1987) and Hammer et al. (2011), is in order to accommodate the needs of breeders and recognition that intergeneric hybrids do not always result in complete amphiploids.
Article H.4.1 of the ICN allows only one correct name for hybrids of known parentage, regardless of genomic composition, stating: “When all the parent taxa can be postulated or are known, a nothotaxon is circumscribed so as to include all individuals recognizably derived from the crossing of representatives of the stated parent taxa (McNeill et al. 2012).” This stipulation attempts to maintain clarity of nomenclature but has the unintended consequence of grouping genomically distinct combinations. An example is ×Triticosecale where there is the possibility of hybridizing diploid or tetraploid rye with wheat, achieving multiple genomic constitutions with only one proper name.
Aligning taxonomy and nomenclature for both wild and directed hybrids has been a source of debate from the beginning of biological classification (Frodin 2004). This distinction is further complicated by cases such as ×Triticosecale where the goal is not simply a novel combination but an evolving, interbreeding crop. The same is true for developing perennial wheat where most of the species used in hybridization are polyploids, yielding a range of potential genomic compositions in their progeny.
One option would be to include genomic composition to allow for recognition of specific combinations at different ploidy levels, or combinations of genomes, at the species level. This would be a more functional grouping for breeding efforts and is perhaps not unreasonable for the production of directed hybrids.
×Tritipyrum aaseae
We propose to elevate the construct tritipyrum of King et al. (1997) to a nothogeneric designation for combinations of species of Triticum with species of Thinopyrum as ×Tritipyrum, in accordance with the ICN (McNeill et al. 2012, Article H.6.2). Nothospecific combinations within this nothogenus can further clarify parentage and this level of definition is valuable to breeders in determining the background of material from other programs (McNeill et al. 2012, Article H.11).
From our work, we would like to offer definition to the new nothospecies ×Tritipyrum aaseae as the combination of T. aestivum and Th. ponticum (Podp.) Barkworth et D.R. Dewey [syn. Thinopyrum elongatum (Host) D.R. Dewey, Elytrigia pontica (Podp.) Holub, Elymus elongatus subsp. ponticus (Podp.) Melderis, Lophopyrum ponticum (Podp.) Á. Löve, Elymus elongatus var. ponticus (Podp.) Dorn, Agropyron elongatum subsp. ponticum (Podp.) Senghas, Elytrigia elongata subsp. pontica (Podp.) Gamisans, Elymus ponticus (Podp.) N. Snow]. There are many synonyms of Th. ponticum based on different classification systems, our choice reflects the importance of genomic as well as morphological characterization for placement (Barkworth and Dewey 1985). Thinopyrum ponticum is an autoallodecaploid with two closely related genomes which will pair autosyndetically in hybrids with T. aestivum (Cai and Jones 1997).
The species name is in honor of Dr. Hannah Aase, a pioneering cytogeneticist who conducted early work on cereal crops at Washington State University (Aase 1926). This recognition reflects her research on intergeneric combinations and suggestion that “Avoidance of further confusion in nomenclature merits a close cooperation of both cyto-geneticists and taxonomists in naming newly obtained aberrants, spontaneous or induced, that may become destined to exist as permanent species (Aase 1946).” A description of a cultivar from this species is offered as an example of additional information that can be included at the variety level to aid breeding efforts. × Tritipyrum Curwen-McAdams et al. nothogen. nov. [Triticum L. ×Thinopyrum Á. Löve]. × Tritipyrum aaseae Curwen-McAdams et al. nothosp. nova. Type: United States. Cultivated by C. Curwen-McAdams in Mount Vernon, Washington, USA. Seed deposited in United States Department of Agriculture Germplasm Resources Information Network (GRIN) PI 676253. [Triticum aestivum L. × Thinopyrum ponticum (Podp.) Barkworth et D.R. Dewey]. Holotype: Washington State University Marion Ownbey Herbarium Accession # 391222. Diagnosis: Caespitose, stems erect, sheath split, auricles stubby, ligules membrane-like. Inflorescence is an erect, lax, fusiform spike, peduncle glabrous, cleistogamous, caryopsis narrow, ovate with a deep ventral crease. Leaves narrow, linear, coarse, with raised parallel veins, hirsute with a deep mid-rib. Plants display an indeterminate flowering habit where new reproductive tillers are continually initiated from the crown. In the greenhouse and climates with mild winters individual plants will display a polycarpic habit, setting seed for two or more seasons.
Etymology: Named in honor of Dr. Hannah Aase, a cytogeneticist at Washington State University who worked with intergeneric hybrids of the Triticeae.
Breeding
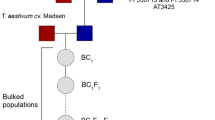
Breeding work conducted at Washington State University over the last 20 years has yielded diverse ×Tritipyrum aaseae combinations with different genetic backgrounds. These lines display a range of phenotypes and are stable at 56 chromosomes with 42 coming from the wheat parent and 14 from Th. ponticum. The original crosses were made using Th. ponticum as the male and T. aestivum ‘Chinese Spring’ as the female. Subsequent generations were backcrossed to T. aestivum ‘Madsen’ and selected for agronomic traits as well as a polycarpic habit under field and greenhouse conditions.
Early generations were genomically unstable, but highly fertile lines have been selected and identified with 56 chromosomes. This number represents the basic allohexaploid genome of T. aestivum and 14 chromosomes from Th. ponticum likely comprised of a hybrid genome as described by Fedak and Han (2005) in other crosses of T. aestivum with Th. ponticum and Th. intermedium (Host.) Barkworth et D.R. Dewey. For breeding purposes, the proportion and origin of these additional chromosomes is important for pairing and recombination at meiosis.
The genomic complexity of intergeneric hybrids means that a range of compositions are possible in the as stable progeny, including chromosomal rearrangements, substitutions and translocations. Visualization of a sample of Thinopyrum accessions using mcGISH showed variation for genomic composition, which will influence what can be inherited through crossing (Kruppa and Molnár-Láng 2016). Breeding work will rely on being able to select lines that are stable and compatible for crossing and selection, which is facilitated by taxonomic grouping based on parentage.
Fluorescent genomic in situ hybridization
Phenotypically stable lines were identified in the field that displayed post-sexual cycle regrowth as described by Lammer et al. (2004). One of those lines, ‘Salish Blue’, was investigated using FGISH in order to determine genomic composition.
Three-day old root meristem tissue was pre-fixed for 24 h at 0.1 °C, then fixed for 48 h in 3:1 ethanol:acetic acid and squashed under a cover glass. Slides were frozen at −80 °C then dehydrated in 45 % acetic acid and 95 % ethanol and chromosomal DNA was denatured by incubation in 70 % formamide at 70 °C for 2 min. DNA probe was prepared using 1 µg of genomic DNA, extracted from leaf tissue of Th. ponticum (USDA GRIN PI 383583), and the BioNick nick translation system (Invitrogen). Blocking DNA was prepared by autoclaving Chinese Spring genomic DNA till it sheared to length of 200–500 nt. A hybridization solution of 25 % dextran sulfate, 60 % formamide, 0.2X SSC, 50 ng of biotinylated probe and 2.5 μg of blocking DNA was applied to each slide and incubated for 16 h at 37 °C. Excess and non-specifically bound probe was removed through a rinse in 2X SSC and incubation at 42 °C for 5 min in 35 % formamide. Slides were incubated in a solution of avidin-fluorescin, followed by biotinylated anti-avidin and a second incubation in avidin-fluorescein. Slides were washed thrice with 4XSSC-Tween20 between treatments. Preparations were counterstained with 10 μg/mL propidium iodide.
Specimens were examined on a Zeiss Axioplan fluorescent microscope at 400X total magnification. The labeled probe generated intense signal on 14 of the 56 chromosomes, distinguishing their Thinopyrum origin. Six replicates generated the same result, indicating stability in the line, and positive fluorescence on 14 Thinopyrum chromosomes of AgCS, a known amphiploid of T. aestivum and Th. elongatum, validated the specificity of the probe (Dvořák and Knott 1974).
Description of ×Tritipyrum aaseae ‘Salish Blue’
Plants caespitose, stems erect, reaching a height of 1.5–2 m or greater at maturity. Sheath split, auricles stubby, ligules membrane-like. Inflorescence is an erect, lax, awnless, fusiform spike, peduncle glabrous, spikelets contain three fertile and two sterile florets, cleistogamous, seed is a blue–green caryopsis, narrow, ovate with a deep ventral crease, anthers yellow, lemma awnless. Glumes half the length of the floret and white or red at maturity, seeds free threshing and rachis tough. Leaves narrow, linear, coarse, with raised parallel veins, hirsute with a deep mid-rib. Plants display an indeterminate flowering habit where new reproductive tillers are continually initiated from the crown. In the greenhouse and climates with mild winters individual plants will display a polycarpic habit, setting seed for two or more seasons.
Plants are intermediate in morphology in most ways between the wheat and wheatgrass parents. The defining characteristics are the ability to initiate post-sexual cycle regrowth, lax heads, coarse leaves and stability at 56 chromosomes, 42 derived from T. aestivum and 14 from Th. ponticum. Figure 1 shows the morphology of the specimen and FGISH image of chromosomes.
Morphology and FGISH of ×Tritipyrum aaseae ‘Salish Blue’. a FGISH image showing 42 chromosomes of wheat and 14 from Thinopyrum. b Comparison of seeds (left to right) of Thinopyrum, ×Tritipyrum aaseae and T. aestivum. c. Flower of ×Tritipyrum aaseae. d Spike of ×Tritipyrum aaseae. e ×Tritipyrum aaseae in the field post-harvest. f Ligule and leaves with coarse venation of ×Tritipyrum aaseae. g New tillers with roots emerging from senesced tiller of ×Tritipyrum aaseae
Discussion
Without taxonomic treatment this crop faces “extinction by nomenclature” (Goncharov 2011). Classifying new combinations as they are created and stabilized develops a base for future researchers to reference, even as the taxonomy of the Triticeae changes in the future. Organizing stable varieties with a nothospecific designation facilitates breeding and taxonomy while keeping valuable information about the origin of materials closely associated and defines the crop for interbreeding.
Reorganization of the Triticeae based on genomic constitution has led to splitting and recombining of many genera (Dewey 1983). While this work has been invaluable for understanding the relationships between different species, the shifting nature of the genomic classifications has highlighted the close homoeology between many of these genomes and the difficulty of making definitive categories (Barkworth et al. 2009). Breeders can operate in this ambiguity, in many cases the homoeology is advantageous, and by defining their combinations they can contribute to the taxonomic conversation as well.
Lack of specificity in nomenclature hinders the development of perennial wheat as a crop. Codifying combinations and describing stable lines allows for searching both the literature and germplasm resources in a way not currently possible. The goal of the breeding effort is to create a new crop type, and accomplishing this requires proper taxonomic placement. The close relationship of many species with Triticum makes the development of diverse combinations possible. Widespread use of stable amphiploids will be hindered without the ability to accurately classify their composition. A commonly agreed on system helps to differentiate the diversity of combinations that researchers use in approaching the common goal.
The potential for chromosomal rearrangements and hybrid genomes in this species, and others generated through wide hybridization, complicates full characterization of individuals and breeding. The nothogenus and nothospecies we have described here are within the articles of the ICN, but the range of possibilities exceeds the current limitation of one correct name for hybrids of species. One possibility is to use genomic information to characterize species when it is a known quality, as is the case with developing new crops. Ultimately there is a larger conversation needed between taxonomists and breeders to find a system that can accommodate new combinations “destined to exist as permanent species”.
References
Aase HC (1926) Chromosome numbers in crop plants. Am J Bot 13:367–372. doi:10.1126/science.39.1007.570
Aase HC (1935) Cytology of cereals. Bot Rev 1:467–496. doi:10.1007/BF02861650
Aase HC (1946) Cytology of cereals. II. Bot Rev 12:255–334
Armstrong JM (1936) Hybridization of Triticum and Agropyron: I. Crossing results and description of the first generation hybrids. Can J Res 14:190–202
Arterburn M, Kleinhofs A, Murray TD, Jones SS (2011) Polymorphic nuclear gene sequences indicate a novel genome donor in the polyploid genus Thinopyrum. Hereditas 148:8–27. doi:10.1111/j.1601-5223.2010.02084.x
Banks PM, Xu SJ, Wang RR-C, Larkin PJ (1993) Varying chromosome composition of 56-chromosome wheat x Thinopyrum intermedium partial amphiploids. Genome 36:207–215
Barkworth ME (1992) Taxonomy of the Triticeae: a historical perspective. Hereditas 116:1–14
Barkworth ME, Dewey DR (1985) Genomically based genera in the perennial Triticeae of North America: identification and membership. Am J Bot 72:767–776
Barkworth ME, Cutler DR, Rollo JS et al (2009) Morphological identification of genomic genera in the Triticeae. Breed Sci 59:561–570. doi:10.1270/jsbbs.59.561
Baum BR (1971) The taxonomic and cytogenetic implications of the problem of naming amphiploids of Triticum and Secale. Euphytica 20:302–306
Baum BR, Gupta PK (1990) Taxonomic examination of Triticale (x Triticosecale). Can J Bot 68:1889–1893
Bernhardt N (2015) Taxonomic treatments of Triticeae and the wheat genus Triticum. In: Molnár-Láng M, Ceoloni C, Doležel J (eds) Alien introgression in wheat: cytogenetics, molecular biology, and genomics. Springer International Publishing, Switzerland, pp 1–19
Brickell CD, Alexander C, David JC et al (2009) International Code of Nomenclature for Cultivated Plants. Scr Hortic 184
Cai X, Jones S (1997) Direct evidence for high level of autosyndetic pairing in hybrids of Thinopyrum intermedium and Th. ponticum with Triticum aestivum. Theor Appl Genet 95:568–572. doi:10.1007/s001220050597
Cai X, Jones SS, Murray TD (2001) Molecular cytogenetic characterization of Thinopyrum genomes conferring perennial growth habit in wheat-Thinopyrum amphiploids. Plant Breed 120:21–26
Ciferri R, Giacomini V (1950) Nomenclator florae Italicae; seu, Plantae vasculares in Italia sponte nascentes, advenae, aut saepius cultae. Ex Typis C., Busca, Italy
Cox TS, Bender M, Picone C et al (2002) Breeding perennial grain crops. CRC Crit Rev Plant Sci 21:59–91. doi:10.1080/0735-260291044188
Dewey DR (1983) Historical and current taxonomic perspectives of Agropyron, Elymus, and related genera. Crop Sci 23:637. doi:10.2135/cropsci1983.0011183X002300040009x
Dvořák J (1976) The cytogenetic structure of a 56-chromosome derivative from a cross between Triticum aestivum and Agropyron elongatum (2 n = 70). Can J Genet Cytol 18:271–279
Dvořák J, Knott DR (1974) Disomic and ditelosomic additions of diploid Agropyron elongatum chromosomes to Triticum aestivum. Can J Genet Cytol 16:399–417
Fedak G, Han FP (2005) Characterization of derivatives from wheat-Thinopyrum wide crosses. Cytogenet Genome Res 109:360–367
Feuillet C, Muehlbauer GJ (eds) (2009) Genetics and genomics of the Triticeae. Springer, New York
Franke R, Nestrowicz R, Senula A, Staat B (1992) Intergeneric hybrids between Triticum aestivum L. and wild Triticeae. Hereditas 116:225–231
Frodin DG (2004) History and concepts of big plant genera. Taxon 53:753–776
Goncharov NP (2011) Genus Triticum L. taxonomy: the present and the future. Plant Syst Evol 295:1–11. doi:10.1007/s00606-011-0480-9
Gupta PK, Baum BR (1986) Nomenclature and related taxonomic issues in wheats, triticales and some of their wild relatives. Taxon 35:144–149
Gupta PK, Fedak G (1986) Hybrids of bread wheat (Triticum aestivum) with Thinopyrum scirpeum (4x) and Thinopyrum junceum (6x). Plant Breed 97:107–111
Hammer K, Filatenko AA, Pistrick K (2011) Taxonomic remarks on Triticum L. and ×Triticosecale Wittm. Genet Resour Crop Evol 58:3–10. doi:10.1007/s10722-010-9590-4
Hayes RC, Newell MT, DeHaan LR et al (2012) Perennial cereal crops: an initial evaluation of wheat derivatives. Field Crops Res 133:68–89. doi:10.1016/j.fcr.2012.03.014
Hsiao C, Chatterton NJ, Asay KH, Jensen KB (1995) Phylogenetic relationships of the monogenomic species of the wheat tribe, Triticeae (Poaceae), inferred from nuclear rDNA (internal transcribed spacer) sequences. Genome 38:211–223. doi:10.1139/g95-026
Jauhar PP, Peterson TS (2013) Synthesis and characterization of advanced durum wheat hybrids and addition lines with Thinopyrum chromosomes. J Hered 104:428–436. doi:10.1093/jhered/ess143
Johnson LPV (1938) Hybridization of Triticum and agropyron: IV. Further crossing results and studies on the F1 hybrids. Can J Res 16:417–444
Kellogg EA (2015) VI. Subfamily Pooideae Benth. (1861) Festucoideae Link (1827). In: Flowering Plants. Monocots: Poaceae. pp 199–265
King IP, Law CN, Cant KA et al (1997) Tritipyrum, a potential new salt-tolerant cereal. Plant Breed 116:127–132
Kishii M, Wang RR-C, Tsujimoto H (2005) GISH analysis revealed new aspect of genomic constitution of Thinopyrum intermedium. In: 5th International Triticeae Symposium p 21
Kruppa K, Molnár-Láng M (2016) Simultaneous visualization of different genomes (J, JSt and St) in a Thinopyrum intermedium ×Thinopyrum ponticum synthetic hybrid (Poaceae) and in its parental species by multicolour genomic in situ hybridization (mcGISH). Comp Cytogenet 10:283–293. doi:10.3897/CompCytogen.v10i2.7305
Lammer D, Cai X, Arterburn M et al (2004) A single chromosome addition from Thinopyrum elongatum confers a polycarpic, perennial habit to annual wheat. J Exp Bot 55:1715–1720. doi:10.1093/jxb/erh209
Larson SR, Kishii M, Tsujimoto H et al (2012) Leymus EST linkage maps identify 4NsL–5NsL reciprocal translocation, wheat-Leymus chromosome introgressions, and functionally important gene loci. Theor Appl Genet 124:189–206. doi:10.1007/s00122-011-1698-1
Li X-M, Lee BS, Mammadov AC et al (2007) CAPS markers specific to Eb, Ee, and R genomes in the tribe Triticeae. Genome 50:400–411. doi:10.1139/g07-025
Liu Z-W, Wang RR-C (1993) Genome constitutions of Thinopyrum curvifolium, T. scirpeum, T. distichum, and T. junceum (Triticeae: Gramineae). Genome 36:641–651
Löve Á (1984) Conspectus of the Triticeae. Feddes Repert 95:425–521
Love RM, Suneson CA (1945) Cytogenetics of certain Triticum-Agropyron hybrids and their fertile derivatives. Am J Bot 32:451–456
Luo M, Dubcovsky J, Dvořák J (1996) Recognition of homeology by the wheat Ph1 locus. Genetics 144:1195–1203
McNeill J, Barrie FR, Buck WR et al (2012) International code of nomenclature for algae, fungi, and plants (Melbourne Code). Regnum Veg 154:1–140
Mujeeb-Kazi A, Hettel GP (eds) (1995) Utilizing wild grass biodiversity in wheat improvement: 15 years of wide cross research at CIMMYT. CIMMYT, Mexico
Mujeeb-Kazi A, Roldan S, Suh DY et al (1987) Production and cytogenetic analysis of hybrids between Triticum aestivum and some caespitose Agropyron species. Genome 29:537–553
Peto FH (1936) Hybridization of Triticum and Agropyron: II. Cytology of the male parents and F1 generation. Can J Res 14:203–214
Sando WJ (1960) Inter- and intra-generic hybrids: an exhibit of crosses between species of Triticum, Secale, Aegilops, and Haynaldia. Crop Res 34:1–15
Sax K (1935) The cytological analysis of species-hybrids. Bot Rev 1:100–117
Scheinost PL, Lammer DL, Cai X et al (2001) Perennial wheat: the development of a sustainable cropping system for the U.S. Pacific Northwest. Am J Altern Agric 16:147–151. doi:10.1017/S0889189300009115
Sears ER (1976) Genetic control of chromosome pairing in wheat. Annu Rev Genet 10:31–51
Sharma HC, Gill BS (1983) Current status of wide hybridization in wheat. Euphytica 32:17–31
Smith DC (1942) Intergeneric hybridization of cereals and other grasses. J Agric Res 64:33–47
Soltis PS, Soltis DE (2009) The role of hybridization in plant speciation. Annu Rev Plant Biol 60:561–588. doi:10.1146/annurev.arplant.043008.092039
Spooner DM, Hetterscheid WLA, van den Berg RG, Brandenburg WA (2003) Plant nomenclature and taxonomy: an horticultural and agronomic perspective. In: Janick J (ed) Horticultural Reviews, John Wiley & Sons, Oxford, pp 1–60
Stace CA (1987) Triticale: a case of nomenclatural mistreatment. Taxon 36:445–452
Suneson CA, El Sharkawy A, Hall WE (1963) Progress in 25 years of perennial wheat development. Crop Sci 3:437–439
Tsitsin NV, Lubimova VF (1959) New species and forms of cereals derived from hybridization between wheat and couch grass. Am Nat 93:181–191
Wang RR-C (1992) Genome relationships in the perennial Triticeae based on diploid hybrids and beyond. Hereditas 116:133–136. doi:10.1111/j.1601-5223.1992.tb00217.x
Wang RR-C (2011) Agropyron and Psathyrostachys. Wild Crop Relat Genomic Breed Resour. doi:10.1007/978-3-642-16057-8
White WJ (1940) Intergeneric crosses between Triticum and Agropyron. Sci Agric 21:198–232
Zhang X, Dong Y, Wang RR-C (1996) Characterization of genomes and chromosomes in partial amphiploids of the hybrid Triticum aestivum x Thinopyrum ponticum by in situ hybridization, isozyme analysis, and RAPD. Genome 39:1062–1071
Acknowledgments
The authors would like to thank all past and present members of the Jones Lab for their contributions and Dr. Pat McGuire (University of California, Davis) for reviewing the manuscript.
Funding
This work funded by an Emerging Research Issues Grant from Washington State University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Curwen-McAdams, C., Arterburn, M., Murphy, K. et al. Toward a taxonomic definition of perennial wheat: a new species ×Tritipyrum aaseae described. Genet Resour Crop Evol 64, 1651–1659 (2017). https://doi.org/10.1007/s10722-016-0463-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-016-0463-3