Abstract
Two new species of nematodes associated with crabs are described from New Caledonia. Gammarinema scyllae sp. n. from the gill chambers of Scylla serrata (Forsskål) is characterised by 3–4 mm long body, small outer labial and cephalic sensilla, distinct ocelli, short straight spicules and sub-cylindrical tail. Monhystrium mangrovi sp. n. from the gill chambers and body cavity of mangrove crab Neosarmatium sp. is characterised by 1–1.4 mm long body; outer labial sensilla longer than cephalic sensilla, amphid located at level with posterior stoma chamber, denticles in posterior stoma chamber and five pairs of genital papilla on tail. Phylogenetic relationships of two new species and other nematodes from the family Monhysteridae are analysed based on 18S and partial 28S rDNA sequences.
Similar content being viewed by others
Introduction
The family Monhysteridae is predominantly free-living taxon with over 180 known species (Fonseca & Decraemer, 2008), that includes several small genera that live in close association with other organisms. These include the genera Gammarinema Kinne & Gerlach, 1953, Monhystrium Cobb, 1920, Tripylium Cobb, 1920, Testudinema Abebe et al., 2012 and Odontobius Roussel del Vauzème, 1834, as well as few species from the genus Halomonhystera Andrássy, 2006.
Gammarinema is a rather small genus, containing species associated decapod and peracarid crustaceans found in both limnic and marine environments. There are only eight known species: Gammarinema ampullocauda (Paramonov, 1926) Lorenzen, 1986, G. cambari (Allen, 1933) Osche, 1955, G. gammari Kinne & Gerlach, 1953, G. ligiae Gerlach, 1967, G. cardisomae Riemann, 1968, G. paratelphusi (Farooqui, 1967) Sudhaus, 1974, G. mesidoteae Belogurov, Kulikov & Russkikh, 1978 and G. prilepskyi Tchesunov & Pletnikova, 1986. Of these, Gammarinema ampullocauda was found free-living in a small lake on the Kinburn Split near the Black Sea (Paramonov, 1926); G. cambari inhabits the gill-chambers of Cambarus acuminatus Faxon and Procambarus blandingii (Harlan), two limnetic decapod species found in the USA (Allen, 1933); G. cardisomae is also found in gill chambers but of a different decapod species, Cardisoma guanhumi Latreille, from marine supralittoral habitats in the Caribbean (Riemann, 1968). Another species, G. gammari has been found in several different amphipod species, such as Gammarus locusta (Linnaeus), G. oceanicus Segerstråle, G. salinus Spooner and G. zaddachi Sexton, all marine and living in the Baltic and Bering Seas (Kinne & Gerlach, 1953; Tchesunov & Pletnikova, 1986). Another European species, Gammarinema ligiae was found on an isopod Ligia oceanica (Linnaeus) living in a marine supralittoral habitat in the Baltic and Helgoland (Gerlach, 1967). The sixth species, G. paratelphusi was found in the gill chambers of a limnetic decapod Paratelphusa sp. from Maharashtra, India (Farooqui, 1967). The last two species G. mesidoteae and G. prilepskyi were discovered on an isopod Mesidotea entomon (Linnaeus) from the Pacific coast of Russia and from Barents Sea (Tchesunov & Pletnikova, 1986).
Monhystrium includes only five known species: M. wilsoni (Baylis, 1915) Cobb, 1920, M. transitans Cobb, 1920, M. inquilinus Riemann, 1969, M. brevis Yoshimura, 1990 and M. tenuis Yoshimura, 1990 which are found exclusively in gill chambers of land crabs in different parts of the world. The first species was discovered in 1910 in the gills of a purple land crab Gecarcinus ruricola (Linnaeus) in Jamaica (Baylis, 1915). The second species was described in 1920 by N.A Cobb from the gill chambers of the same purple land crab and subsequently also from the gills of the blackback land crab G. lateralis (Guérin) from Jamaica (Cobb, 1920). In 1969 F. Riemann described the third species of Monhystrium, M. inquilinus; this new species was also found in the gill chambers of a land crab, but this time in a blue land crab Cardisoma guanhumi caught in Colombia (Riemann, 1969). The last two species to be added to the genus were Monhystrium tenuis and M. brevis, described by K. Yoshimura (1990); M. tenuis was found in the gills of Parasesarma plicatum (Latreille), P. pictum (De Haan) and Clistocoeloma merguiense de Man while M. brevis was found in the gill chamber of the red-clawed crab Chiromantes haematocheir (De Haan), C. dehaani (H. Milne Edwards) and Chasmagnathus convexus (De Haan) and also in the gills of flower crabs Orisarma intermedium (De Haan).
Materials and Methods
Sampling and specimen preparation. Nematodes were found during parasitological inspection of land crab specimens obtained from the local inhabitants. Nematodes were carefully removed from the gills of crabs and immediately preserved in 5% formaldehyde solution, 95% ethanol solution and RNAlater. For light microscopy, specimens were transferred to pure glycerine using Seinhorst’s (1959) rapid method as modified by De Grisse (1969). Permanent nematode mounts on glass slides were prepared using the paraffin wax ring method. All curved structures were measured along the curved median line. Terminology follows Maggenti et al. (2005). Abbreviations are according to Hunt & Palomares-Ruis (2012).
Molecular analysis. DNA extraction was performed on two individuals for each species. Individual nematodes were each placed in 1.5 ml microcentrifuge tubes containing 20 μl buffer ATL (Qiagen, Sweden) and stored at -20°C until all samples were ready for extraction. During the extraction, 160 μl of buffer ATL was added to each sample. This was followed by the addition of 20 μl proteinase K, vortexing and incubation in an incubating microplate shaker at 56°C and 300 rpm overnight. The lysed samples were further processed to obtain pure DNA following the manufacturer’s instructions for genomic DNA extraction using the Qiagen QiAmp DNA Micro kit. Two regions of the rDNA gene, the nearly full-length of the 18S and the D2–D3 expansion segment of 28S, were amplified. The approximately 1800 bp region of the 18S rRNA gene was amplified as two overlapping fragments using the primer sets 988F–1912R for the first fragment and 1813F–2646R for the second fragment (Holterman et al., 2006). Polymerase chain reaction (PCR) for both fragments was performed in 25 μl reaction mix using Illustra Hot Start Mix RTG 0.2 ml reaction kit (GE Healthcare Life Sciences, Sweden). The reaction mix consisted of 1 μl (0.4 μM) of each primer, 2 μl template DNA and 21 μl nuclease-free water. The reaction conditions were 5 min at 95°C; 5 cycles of (30 sec at 94°C, 30 sec at 45°C and 30 sec at 72°C); 35 cycles of (30 sec at 94°C, 30 sec at 54°C and 30 sec at 72°C); and a final extension for 5 min at 72°C. The D2–D3 segment of the 28S rRNA gene was amplified using the primers D2Af and D3Br (Nunn, 1992). PCR was performed in 25 μl reaction mix containing 1 μl (0.4 μM) of each primer, 2 μl template DNA and 21 μl nuclease-free water. The PCR conditions were 4 min at 94°C; 35 cycles of (94°C for 60 sec, 54°C for 90 sec and 72°C for 2 min); final extension for 10 min at 72°C. Enzymatic PCR clean-up was performed on the PCR product using Exonuclease I and Shrimp Alkaline Phosphatase (New England Biolabs, MA, USA). The purified PCR products were sent out to Macrogen Europe B.V. (Amsterdam, the Netherlands) for sequencing. Each amplicon was sequenced in both directions using the forward and reverse PCR primers. The trace files of the individual sequences were visualized inside BioEdit (Hall, 1999) and trimmed to high quality. The trimmed forward and reverse sequences were then assembled using Fragment Merger online tool (Bell & Kramvis, 2013). The two fragments of the 18S rRNA gene were also assembled into contigs using the Fragment Merger online tool.
Phylogenetic analysis. Alignment from Ahmed and Holovachov (2020) for 18S rRNA gene was used as template for alignment and annotation. New sequences were aligned to a fixed template alignment using AliView (Larsson, 2014). Partial 28S rDNA sequences were aligned de novo in AliView. Phylogenetic trees were built using RAxML ver. HPC2 (Stamatakis, 2014) via the CIPRES portal (Miller et al., 2010) for the Maximum Likelihood inference of the partitioned dataset. The GTR nucleotide substitution model was used for non-paired sites, whereas the RNA7A (Higgs, 2000) substitution model was used for paired sites. Bootstrap ML analysis was performed using the rapid bootstrapping option with 1000 iterations.
Monhysteridae de Man, 1876
Gammarinema Kinne & Gerlach, 1953
-
Type species:
-
Gammarinema gammari Kinne & Gerlach, 1953, by original designation.
-
Other species:
-
Gammarinema ampullocauda (Paramonov, 1926) Lorenzen, 1986
-
= Monhystera ampullocauda Paramonov, 1926
-
Gammarinema cambari (Allen, 1933) Osche, 1955
-
= Rhabditis cambari Allen, 1933
-
Gammarinema ligiae Gerlach, 1967
-
Gammarinema paratelphusi (Farooqui, 1967) Sudhaus, 1974
-
= Branchinema paratelphusi Farooqui, 1967
-
Gammarinema cardisomae Riemann, 1968
-
Gammarinema mesidoteae Belogurov, Kulikov & Russkikh, 1978
-
Gammarinema prilepskyi Tchesunov & Pletnikova, 1986
-
Gammarinema scyllae sp. n.
Genus diagnosis: Body length medium to long (0.7–4.0 mm). Inner labial sensilla papilliform; outer labial and cephalic sensilla in one circle, same or different in length, with either outer labial or cephalic sensilla being longer than the others. Amphids round; less than 1.5 head diameters from the anterior body end. Ocelli present or absent. Buccal cavity funnel shaped, with weakly to moderately cuticularized anterior chamber of stegostom, and narrow posterior chamber of stegostom; posterior chamber with 3 or 6 denticles or without any. Pharynx cylindrical. Progaster present. Ventral gland well-developed and visible, excretory pore opens along the anterior region of the pharynx or within the labial region. Ovary and testis on the right-hand side of intestine. Spicules simple and narrow, straight to arcuate. Gubernaculum platelike, with or without apophysis. Precloacal supplements may be present. Tail conoid to subcylindrical. Caudal glands opening via common spinneret. Usually found associated with crustaceans.
Gammarinema scyllae sp. n.
Type host and locality: The nematodes were found in the gill chambers of the mud crab Scylla serrata (Forskål) collected near Wagap, Poindimié commune, Northern Province, New Caledonia (HYNC4666; July 12, 2018).
Type material: Holotype male and two males and seven females paratypes on slides MNHN-BN514–MNHN-BN518 are deposited in the meiofauna collection of the National Museum of Natural History in Paris, France (MNHN). Seven males, six females and 73 juveniles paratypes on slides SMNH Type-9353–SMNH Type-9359 are deposited in the Invertebrate type collection of the Department of Zoology, Swedish Museum of Natural History, Stockholm, Sweden.
Etymology: The species name scyllae is derived from its host name Scylla.
ZooBank registration: urn:lsid:zoobank.org:pub:A0F29FA2-B06B-4CFF-A100-BE4F292E8188 (publication); urn:lsid:zoobank.org:act:CAA34FB3-AD20-43B8-B3B6-73D1A64914F9 (species).
GenBank acc. numbers: Sequences obtained are deposited in GenBank under the accession numbers MZ274175 and MZ274176 for the D2-D3 segment of the 28S rRNA gene and MZ274171 and MZ274172 for the nearly full-length 18S rRNA gene.
Description
Diagnosis: Gammarinema scyllae sp. n. is characterised by 3–4 mm long body, small outer labial and cephalic sensilla, distinct ocelli, short straight spicules and sub-cylindrical tail. In gill chambers of the mud crab Scylla serrata.
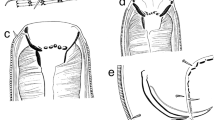
Adult. (Figures 1, 2, 3, Table 1). Body cylindrical, ventrally curved upon fixation in females and in males, tapering slightly towards both extremities along pharyngeal region and on tail. Cuticle smooth. Somatic sensilla present, small setiform. Body pores absent. Lateral alae absent. Cephalic region flattened. Six equal lips surrounding mouth opening. Inner labial sensilla small, located on anterior surface of lips. Outer labial sensilla small setiform, located at the base of lip region. Cephalic sensilla papilliform, located at the same level as labial sensilla. Amphideal opening round, located at level of stoma base. Ocelli present. Buccal cavity conoid in general, cheilostom and gymnostom both cylindrical and very short; stegostom funnel-shaped (conoid anteriorly and tubular posteriorly), with weakly cuticularised walls. Posterior chamber poorly defined, denticles indistinct, edges of pharyngeal radii visible. Pharynx uniformly muscularized along its entire length, gradually widening posteriorly but without any valves or bulbs. Cardia small and conical. Intestinal lumen well developed, progaster present. Secretory-excretory system and secretory-excretory pore present, renette cell located along anterior part of intestine, excretory pore located about 1.2 times labial region diameter from the anterior end. Tail cylindrical, with broadly rounded terminus. Caudal glands present, opening towards extension via a common spinneret. Caudal gland cells located in the tail. Spinneret not cuticularised.
Female. Reproductive system monodelphic. Ovary branch outstretched, extends anterior but not reaching the cardia, on the right-hand side of intestine. Post-vulval uterine sac absent. Vagina directed anteriorly. Vulva located posterior to midbody, a transverse ventral slit, not cuticularized.
Male. Reproductive system monorchic, on the right-hand side of intestine. Spicules paired and symmetrical, straight and relatively thin conoid, equal to 0.7–0.8 anal body diameters in length. No pre- or post- cloacal sensilla or supplements.
rRNA. Sequences include two nearly full length 18S rRNA gene and two partial 28S rRNA gene representing D2D3 domain. Sequence variability of the 18S rRNA gene was small, less 1–7 bases difference; sequences of D2D3 domain of 28S rRNA gene were identical. This is the first species from the genus Gammarinema to be sequenced.
Relationships
None of the previously described species was sequenced, therefore the differentiation of new species is based on morphological characters (see also Table 2), host and geographic distribution.
Gammarinema ampullocauda is smaller than G. scyllae sp. n. with body length of 1.88–2.16 mm compared to 2.97–3.74 mm in G. scyllae sp. n. The a-value of the G. ampullocauda is 46–48 and in G. scyllae sp. n. it is 55–76. The tail is shorter in G. ampullocauda (175–180 µm) than in G. scyllae sp. n. (201–270 µm). The length of cephalic sensilla is 6 µm in G. ampullocauda compared to 1.5–3.5 µm in G. scyllae sp. n. Spicules are shorter in G. ampullocauda (25 µm) than in G. scyllae sp. n. (32–36 µm). G. ampullocauda was found in Black Sea, its host is unknown.
Gammarinema cambari is described very superficially. The new species is much bigger than G. cambari with body length reaching 2.97–3.74 mm in new species compared to 0.77 mm in G. cambari. The shape of the tail differs between two species, the tail of G. scyllae sp. n. is sub-cylindrical and the tail of G. cambari is conoid. The spicules are arcuate in G. cambari but straight in G. scyllae sp. n. G. cambari was found in North Carolina in the USA and uses Cambarus acuminatus and C. blandingii as a host, while G. scyllae sp. n. was found in New Caledonia.
Gammarinema scyllae sp. n. is more than twice as big as G. cardisomae that has body length of 1.12–1.16 mm compared to 2.97–3.74 mm in G. scyllae sp. n. Moreover, G. scyllae sp. n. has higher values of many ratios than G. cardisomae: a-value is 44–50 in G. scyllae sp. n. compared to 55–76 in G. cardisomae, b-value is 5.8–6.0 in G. scyllae sp. n. compared to 7.1–8.7 in G. cardisomae and c-value is 8.8–9.1 in G. scyllae sp. n. compared to 10.8–15.1 in G. cardisomae. Exception is a c´-value where G. cardisomae has higher value of 6.0–7.9 compared to 4.7–6.3 in G. scyllae sp. n. The tails of the two species are the same in shape but differ in length: 128 µm in G. cardisomae and 201–270 µm in G. scyllae sp. n. G. cardisomae has three teeth while teeth are absent in G. scyllae sp. n. The spicules are different in shape in the two species, the spicules in G. cardisomae are arcuate and in G. scyllae sp. n. the spicules are straight. G. cardisomae has a gubernaculum with apophysis which is absent in the G. scyllae sp. n. G. cardisomae was found in Colombia, South America.
Gammarinema scyllae sp. n. is somewhat bigger than G. gammari with a body length ranging from 1.62–2.99 mm in the latter and 2.97–3.74 mm in the former. The a-, b- c- ratios are similar/overlapping, but the c´-value is different, 6–10 in the G. gammari compared to 4.7–6.3 in G. scyllae sp. n. The amphid is located 9–12 µm from the anterior end in G. gammari compared to 6–8 µm in G. scyllae sp. n. G. gammari has six small teeth while teeth are absent in the G. scyllae sp. n. G. gammari has slightly larger (39–57 µm) and weakly arcuate spicules while G. scyllae sp. n. has shorter (32–36 µm) and nearly straight spicules. G. gammari has a small plate-like gubernaculum, whereas gubernaculum is absent in G. scyllae sp. n. G. gammari uses multiple species of Gammarus as host and is found in Northern Europe.
Gammarinema ligiae is smaller with a body length of only 0.87–1.18 mm while G. scyllae sp. n. is 2.97–3.74 mm long. Body proportions are also different: a-value is 28–43 in G. ligiae and 55–76 in G. scyllae sp. n., b-value is 5.8–7.0 in G. ligiae and 7.1–8.7 in G. scyllae sp. n., c-value is 8.3–10.8 in G. ligiae and 10.8–15.1 in G. scyllae sp. n. The tail is shorter (90–130 µm) in G. ligiae than in G. scyllae sp. n. (201–270 µm). The amphid in G. ligiae is positioned more posterior from the anterior end (13 µm) than in G. scyllae sp. n. (6–8 µm). G. ligiae has six small teeth that G. scyllae sp. n. lacks. The spicules are straight and slightly larger (32–36 µm) in G. scyllae sp. n. while they are weakly arcuate and smaller (28–31 µm) in G. ligiae. The gubernaculum is present and has apophysis in G. ligiae but is absent in G. scyllae sp. n. G. ligiae stands out from other species of the genus by having cuticularised vulva. G. ligiae is distributed in Northern Europe (Helgoland and Kiel Bay) and uses Ligia oceanica as a host.
Gammarinema scyllae sp. n. is bigger than G. mesidoteae with a body length of 2.97–3.74 mm compared to 2.31–2.95 mm in G. mesidoteae. The cephalic sensilla are slightly longer (4–5 µm) in G. mesidoteae than in G. scyllae sp. n. (1.5–3.5). The amphid is positioned 10–13 µm from the anterior end in G. mesidoteae and 6–8 µm in G. scyllae sp. n. The spicules in G. mesidoteae are both longer 36–39 µm and have a different shape (weakly arcuate) compared to G. scyllae sp. n. that has 32–36 µm long straight spicules. G. mesidoteae has gubernaculum in the shape of a small plate while gubernaculum is completely absent in G. scyllae sp. n. G. mesidoteae lives in Far East and is known to have Mesidotea entomon as a host.
Gammarinema paratelphusi is described very superficially, with few measurements given, and with some unusual morphological features, such as cephalic sensilla arranged in pairs or pre- and postcloacal sensilla present in males. The new species can be easily separated from G. paratelphusi in having shorter (32–36 µm) and differently shaped (straight) spicules, which are weakly arcuate and 55–58 µm long in G. paratelphusi, absence of teeth in stoma (present in G. paratelphusi), absence of pre- and postcloacal sensilla in male (present in G. paratelphusi). G. paratelphusi was found in India in a freshwater crab Paratelphusa sp.
Gammarinema prilepskyi has 2.4 mm long body while G. scyllae sp. n. is 2.97–3.74 mm long. The a-value differs between the two species, 35–48 in G. prilepskyi and 55–76 in G. scyllae sp. n. Same with c-value which is 16.1–22.1 in G. prilepskyi and 10.8–15.1 in G. scyllae sp. n. The tail is cylindrical with digitate tip in G. prilepskyi and sub-cylindrical in G. scyllae sp. n. The cephalic sensilla are longer in G. prilepskyi (6–9 µm) than in G. scyllae sp. n. (1.5–3.5 µm). The length and shape of spicules are different with G. prilepskyi having weakly arcuate 52 µm long spicules, while the spicules in G. scyllae sp. n. are straight and 32–36 µm long. G. prilepskyi has small plate-like gubernaculum and G. scyllae sp. n. has none. The geographic distribution of G. prilepskyi is limited to the Barents Sea and the host is Mesidotea entomon.
Identification key to species of the genus Gammarinema
-
1. Ocelli present ... G. scyllae sp. n.
-
– Ocelli absent ... 2
-
2. Male with numerous (15–20) supplements ... G. cardisomae
-
– Male without supplements ... 3
-
3. Labial sensilla grouped in pairs ... G. paratelphusi
-
– Labial sensilla equidistantly arranged ... 4
-
4. Body shorter than 0.8 mm; tail conoid; spicules equal to two cloacal body diameters in length ... G. cambari
-
– Body longer than 0.8 mm; tail subcylindrical; spicules less than 1.5 cloacal body diameters in length... 5
-
5. Tail with distinct digitate distal part; outer labial and cephalic sensilla equal in length ... G. prilepskyi
-
– Tail without digitate distal part; outer labial and cephalic sensilla unequal in length... 6
-
6. Vulva distinctly cuticularized; gubernaculum with apophysis ... 7
-
– Vulva not cuticularized; gubernaculum plate-like or absent ... 8
-
7. Outer labial sensilla longer than cephalic sensilla; tail relatively long (210-270 µm; c’=6–10) ... G. gammari
-
– Cephalic sensilla longer than outer labial sensilla; tail relatively long (90-130 µm; c’=4–6) ... G. ligiae
-
8. Spicules 25 µm long ... G. ampullocauda
-
– Spicules 36–39 µm long ... G. mesidoteae
Monhystrium Cobb, 1920
-
Type species:
-
Monhystrium transitans Cobb, 1920, by original designation.
-
Other species:
-
Monhystrium wilsoni (Baylis, 1915) Cobb, 1920
-
= Monhystera wilsoni Baylis, 1915
-
Monhystrium inquilinus Riemann, 1969
-
Monhystrium brevis Yoshimura, 1990
-
Monhystrium tenuis Yoshimura, 1990
-
Monhystrium mangrovi sp. n.
Genus diagnosis: Body length medium to long (0.8–1.7 mm). Inner labial sensilla papilliform; outer labial and cephalic sensilla in one circle, same or different in length, with either outer labial or cephalic sensilla being longer than the others. Amphids round; less than 1.5 head diameters from the anterior body end. Ocelli present or absent. Buccal cavity cuticularized and distinctly divided into two chambers: anterior chamber conoid; posterior chamber spherical, with sharp tooth-like anterior edges, with or withour denticles. Pharynx cylindrical. Progaster present. Ventral gland present or absent, excretory pore opens along the anterior region of the pharynx. Ovary and testis on the right-hand side of intestine. Spicules simple and narrow, weakly arcuate. Gubernaculum platelike. Precloacal spine may be present. Bursa present. Paired subventral papilla present, pre- and postcloacal, within or outside bursa. Tail conoid to subcylindrical, digitate. Caudal glands opening in via common spinneret. Usually found in gill chambers of crustaceans.
Note: The genera Monhystrium and Diplolaimelloides Meyl, 1954 are close morphologically, with Diplolaimelloides delyi Andrássy, 1958 being also found in the gill chambers of land crabs, the only clear morphological difference between them is the more strongly developed and cuticularized posterior stoma chamber with inward-pointing tooth-like anterior edges in Monhystrium. Sequenced species from two genera do not form a monophyletic lineage (Figure 7), however, the limited taxon sampling seriously undermines our understanding of the phylogeny of this group in general.
Monhystrium mangrovi sp. n.
Type host and locality: The nematodes were found in the gill chambers of the mangrove crab Neosarmatium sp. collected near Poya, Poya commune, west coast of the Northern Province, New Caledonia (HYNC4625; July 3, 2018).
Type material: Holotype male and eleven juvenile paratypes on slides MNHN-BN512 and MNHN-BN13 are deposited in the meiofauna collection of the National Museum of Natural History in Paris, France (MNHN). Three females and 12 male paratypes on slides SMNH Type-9351 and SMNH Type-9352 are deposited in the Invertebrate type collection of the Department of Zoology, Swedish Museum of Natural History, Stockholm, Sweden.
Etymology: The species name mangrovi refers to the host of this species, a species of a mangrove crab.
ZooBank registration: urn:lsid:zoobank.org:pub:A0F29FA2-B06B-4CFF-A100-BE4F292E8188 (publication); urn:lsid:zoobank.org:act:B8A75B36-6BB5-4C72-88B2-EBDC03B6AC5D (species).
GenBank acc. numbers: Sequences obtained are deposited in GenBank under the accession numbers MZ274177 and MZ274178 for the D2-D3 segment of the 28S rRNA gene and MZ274173 and MZ274174 for the nearly full-length 18S rRNA gene.
Description
Diagnosis: Monhystrium mangrovi sp. n. is characterised by 1–1.4 mm long body; outer labial sensilla longer than cephalic sensilla, amphid located at level with posterior stoma chamber, denticles in posterior stoma chamber and five pairs of genital papilla on tail. In gill chambers and body cavity of mangrove crab Neosarmatium sp.
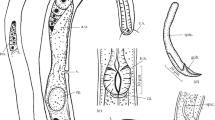
Adult. (Figures 4, 5, 6, Table 3). Body cylindrical, nearly straight upon fixation, tapering slightly towards both extremities along pharyngeal region and on tail. Cuticle smooth. Somatic sensilla present, small setiform. Body pores absent. Lateral alae absent. Cephalic region rounded. Six equal lips surrounding mouth opening. Inner labial sensilla papilliform, located on anterior surface of lips. Outer labial sensilla setiform, located at the base of lip region. Cephalic sensilla small papilliform, located at the same level as outer labial sensilla. Amphideal opening round, located at level of posterior stoma chamber. Ocelli absent. Buccal cavity composed of two chambers. Anterior chamber of buccal cavity conical. Posterior chamber broad spherical with multiple teeth on the anterior part of its ventral surface. Pharynx uniformly muscularized along its entire length, gradually widening posteriorly but without any valves or bulbs. Cardia small. Intestinal lumen well developed, progaster present. Secretory-excretory system and secretory-excretory pore not visible, absent. Tail conical, with digitiform terminus. Caudal glands present, opening towards exterior via a common spinneret. Caudal gland cells located in the tail. Spinneret not cuticularised.
Monhystrium mangrovi sp. n. a. Anterior end (combined view); b. Vulval region; c. Male posterior end, ventral view (arrowheads point to genital papilla); d. Male posterior end, lateral view (arrowheads point to genital papilla); e. Female tail; f. Entire male; g. Entire female. Scale bars: a–e = 20 µm, f–g = 500 µm.
Female. Reproductive system monodelphic. Ovary branch outstretched, extends anterior but not reaching the cardia, on the right-hand side of intestine. Post-vulval uterine sac absent. Vagina directed anteriorly. Vulva located posterior to midbody, a transverse ventral slit, not cuticularised.
Male. Reproductive system monorchic, on the right-hand side of intestine. Spicules paired and symmetrical, arcuate, very long, curved ventral, with angular manubrium and cylindrical shaft, thin velum and lateral projections near their tips equal to 2.1–3.0 anal body diameters in length. Gubernaculum plate-like. No midventral pre- or post- cloacal supplements. Precloacal spine present, short distance in front of cloacal opening. Bursa well-developed, starts anterior to cloaca and extends to the posterior third of tail. Five pairs of genital papillae present: one pair just posterior to cloaca, three pairs near posterior end of bursa and one pair just behind bursa
rRNA. Sequences include two nearly full length 18S rRNA gene and two partial 28S rRNA gene representing D2D3 domain. Sequence variability of 18S rRNA gene was small, less than 1–3 bases difference; sequences of D2D3 domain of 28S rRNA gene were identical. This is the first species from the genus Monhystrium to be sequenced.
Relationships
None of the previously described species was sequenced, therefore the differentiation of the new species is based on morphological characters (see also Table 4), host and geographic distribution. The new species can be separated from all other known species of the genus by the presence of denticles in the posterior stoma chamber.
Monhystrium mangrovi sp. n. is smaller than M. wilsoni with body length of 1.09–1.37 mm compared to 1.4–1.7 mm in M. wilsoni. M. wilsoni has longer tail (176 µm) with c´-value of 4.8 compared to 79–129 µm in M. mangrovi sp. n. with c´-value of 2.2–3.4. The position of the amphid is different: at level with anterior stroma chamber in M. wilsoni compared to at level with posterior stoma chamber in M. mangrovi sp. n. There are differences in the number of genital papilla in both species: M. wilsoni has two pairs of precloacal and eight pairs of postcloacal genital papilla within bursa, while M. mangrovi sp. n. have no precloacal papilla and four pairs of postcloacal papilla in bursa arranged in two groups (1+3). Precloacal spine is present in M. wilsoni but absent in M. mangrovi sp. n. The mode of reproduction is viviparous in M. wilsoni and oviparous in M. mangrovi sp. n. M. wilsoni is found in Jamaica.
Monhystrium mangrovi sp. n. is bigger than M. transitans with body length of 1.09–1.37 mm compared to 0.83–0.90 mm in M. transitans. M. transitans has relatively shorter pharynx (b-value 9.4–9.7) compared to M. mangrovi sp. n. (b-value 6.4-8.0). In M. transitans the amphid is located at level with the anterior stoma chamber and in M. mangrovi sp. n. at level with posterior stoma chamber. The M. transitans has one pair of precloacal genital papilla and four pairs of postcloacal genital papilla within bursa while M. mangrovi sp. n. has no precloacal genital papilla in bursa and four pairs of postcloacal papilla arranged in two groups (1+3) in bursa. The known distribution of M. transitans is limited to Jamaica.
Monhystrium aff. transitans is smaller than M. mangrovi sp. n. with a body length of 0.83–1.0 mm compared to 1.09–1.37 mm in M. mangrovi sp. n. The amphid is located at level with anterior stoma chamber in M. aff. transitans but at level with posterior stoma chamber in M. mangrovi sp. n. The spicules are much shorter in M. aff. transitans (31 µm) than in M. mangrovi sp. n. (62–73 µm). Precloacal spine is present in M. aff. transitans but absent in M. mangrovi sp. n. Geographic distribution of M. aff. transitans is limited to Colombia.
Monhystrium inquilinus has shorter tail both in males 60 µm (compared to 79–100 µm in M. mangrovi sp. n.) and in females 86 µm (compared to 112–129 µm in M. mangrovi sp. n.). Spicules are shorter in M. inquilinus (50 µm) compared to 62–73 µm in the new species. Only three pairs of genital papilla are present postcloacal in bursa in M. inquilinus but in M. mangrovi sp. n. there are four pairs of postcloacal papillae arranged in two groups (1+3) in bursa. Precloacal spine, and ocelli are present in M. inquilinus but both structures are absent in M. mangrovi sp. n. M. inquilinus is found in Colombia so far.
Monhystrium tenuis is slimmer than the new species, with a-value being higher (44–48) comparing to M. mangrovi sp. n. (23–32). The length of the spicules is different between species, 47–54 µm in M. tenuis and 62–73 µm in the M. mangrovi sp. n. There are only two pairs of genital papillae postcloacal in bursa in M. tenuis while M. mangrovi sp. n. has four pairs of postcloacal genital papilla arranged in two groups (1+3) in bursa. M. tenuis is found in Japan.
Monhystrium brevis is smaller with a body length of 0.87–1.04 mm compared to 1.09–1.37 mm in M. mangrovi sp. n. The length of the spicules smaller in M. brevis (43–60 µm) than in M. mangrovi sp. n. (62–73 µm). Another thing that separates the two species is that M. mangrovi sp. n. has four pairs of postcloacal genital papilla arranged in two groups (1+3) in bursa and M. brevis has only two pairs. M. brevis is found in Japan.
Identification key to species of the genus Monhystrium
-
1. Posterior stoma chamber with numerous denticles ... M. mangrovi sp. n.
-
– Posterior stoma chamber without denticles ... 2
-
2. Amphid at the level with anterior stoma chamber ... 3
-
– Amphid at the level with posterior stoma chamber ... 4
-
3. Cephalic sensilla papilliform; body 1.4–1.7 mm long ... M. wilsoni
-
– Cephalic sensilla setiform; body 0.8–1.0 mm long ... M. transitans
-
4. Ocelli present; precloacal spine present ... M. inquilinus
-
– Ocelli absent; precloacal spine absent ... 5
-
5. Body 1.2–1.5 mm long; a > 40; c > 12 ... M. tenuis
-
– Body 0.9–1.1 mm long; a < 40; c < 12 ... M. brevis
Phylogenetic position of Gammarinema and Monhystrium
Two genes were used to build two different phylogenetic trees, 18S (Fig. 7) and 28S rDNA (Fig. 8), with members of the family Linhomoeidae being used as outgroups. The phylogeny based on 18S rDNA was the one most reliable with reasonably high bootstrap support values overall. It also covered a broader taxonomic diversity. The 28S rDNA tree included fewer taxa and showed lower bootstrap support for many clades. Both trees suggest Gammarinema scyllae sp. n. is most closely related to Monhystrium mangrovi sp. n., both belonging to the family Monhysteridae. Current phylogeny indicates that both genera, Gammarinema and Monhystrium originate from a recent common ancestor. Diplolaimella and Diplolaimelloides were recovered as the closest relatives to the two genera and the four form a very well supported clade, in agreement with morphology-based theories. Moreover, the fact that all known species of Monhystrium are found exclusively in gill chambers of different land crabs and eight out of nine known species of Gammarinema are associated with crustaceans suggests that the common ancestor was also associated with crustaceans. Future studies should focus on sequencing other species from Gammarinema, Monhystrium and also of closely related commensalistic (Odontobius, Tripylium, Diplolaimelloides delyi) and free-living species to better understand the phylogeny of this group. Although the phylogeny did not support the subfamilies Diplolaimellinae and Monhysterinae, the family Monhysteridae received maximal support.
Availability of data and material
All studied specimens are deposited in permanent and accessible repositories: National Museum of Natural History in Paris, France and Swedish Museum of Natural History, Stockholm, Sweden. Sequences are deposited in GenBank.
Code availability
Not applicable.
References
Ahmed, M., & Holovachov, O. (2020). Description of a new marine predatory nematode Latronema dyngi sp. nov. (Nematoda, Chromadorida, Selachinematidae) from the west coast of Sweden and an updated phylogeny of Chromadoria. Marine Biodiversity, 50, 113. https://doi.org/10.1007/s12526-020-01129-w
Allen, S. A. (1933) Parasites and commensals of North Carolina crayfishes. Journal of the Elisha Mitchell Scientific Society, 49, 119–121.
Andrássy, I. (2006). Halomonhystera, a new genus distinct from Geomonhystera Andrássy, 1981 (Nematoda: Monhysteridae). Meiofauna Marina, 15, 11–24
Baylis, H. A. (1915). Two new species of Monhystera (Nematodes) inhabiting the gill-chambers of land-crabs. Annals and Magazine of Natural History, 16(95), 414–421.
Bell, T., & Kramvis, A. (2013). Fragment merger: An online tool to merge overlapping long sequence fragments. Viruses, 5(3), 824–833.
Cobb, N. A. (1920). One hundred new nemas (type species of 100 new genera). Contributions to a Science of Nematology, 9, 217–343.
De Grisse, A. T. (1969). Redescription ou modification de quelques tech- niques utilisés dans l’études dęs nématodes phytoparaires. Mededelingen van de Rijksfaculteit Landbouwwetenschappen te Gent, 34, 351–369.
Farooqui, M. N. (1967). Branchinema paratelphusi gen. et sp. nov. from a fresh-water crab Paratelphusa sp. Zoologischer Anzeiger, 178, 354–358.
Fonseca, G., & Decraemer, W. (2008). State of the art of the free-living marine Monhysteridae (Nematoda). Journal of the Marine Biological Association of the United Kingdom, 88, 1371–1390.
Gerlach, S. A. (1967). Zwei neue freilebende marine Nematoden vergesellschaftet mit Crustaceen des Supralitorals. Veröffentlichungen des Institut Meeresforschung Bremerhaven, 10, 209–215.
Hall, T. A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symposium series, 41, 95–98.
Higgs, P. G. (2000). RNA secondary structure: Physical and computational aspects. Quarterly Reviews of Biophysics, 33(3), 199–253. https://doi.org/10.1017/s0033583500003620
Holterman, M., van der Wurff, A., van den Elsen, S., van Megen, H., Bongers, T., Holovachov, O., Bakker, J., & Helder, J. (2006). Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown clades. Molecular Biology and Evolution, 23(9), 1792–1800. https://doi.org/10.1093/molbev/msl044
Hunt, D. J., & Palomares-Rius, J. E. (2012). General morphology and morphometries of plant-parasitic nematodes. In R. H. Manzanilla-López, & N. Marbán-Mendoza (Eds.), Practical plant Nematology (pp. 25–64). Mundi-Prensa.
Kinne, O., & Gerlach, S.A. (1953). Ein neuer Nematode als Kommensale auf Brackwassergammariden, Gammarinema gammari n. g. n. sp. (Monhysteridae). Zoologischer Anzeiger, 151, 192–203.
Larsson, A. (2014). AliView: A fast and lightweight alignment viewer and editor for large data sets. Bioinformatics, 30(22), 3276–3278. https://doi.org/10.1093/bioinformatics/btu531
Maggenti, A. B., Maggenti, A. R., & Gardner, S. L. (2005, June 9). Online dictionary of invertebrate zoology. digitalcommons.unl.edu Retrieved June 30, 2021, from https://digitalcommons.unl.edu/onlinedictinvertzoology/
Miller, M. A., Pfeiffer, W., & Schwartz, T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE), 1–8.
Nunn, G. B. 1992. Nematode molecular evolution: An investigation of evolutionary patterns among nematodes based upon DNA sequences. University of Nottingham, UK.
Paramonov, A. A. (1926). Anatomische und Systematische Beiträge zur Kenntniss der Freilebenden Nematoden. Russkii Zoologicheskii Zhurnal, 6, 44–56.
Riemann, F. (1968). Gammarinema cardisomae nov. spec. (Nematoda: Monhysteridae) aus dem Kiemenraum karibisher Landkrabbe. Mitteilungen aus dem Instituto Colombo-Alemán de Investigaciones Científicas Punta de Betín, 2, 39–43.
Riemann, F. (1969). Nematoden aus den Kiemenraum karibischer Landkrabben: Monhystrium inquilius nov. spec. (Monhysteridae). Veröffentlichungen des Institut Meeresforschung Bremerhaven, 11, 239–244.
Riemann, F. (1970). Das Kiemenlückensystem von Krebsen als Lebensraum der Meiofauna, mit Beschreibung freilebender Nematoden aus karibischen amphibisch lebenden Decapoden. Veröffentlichungen des Institut Meeresforschung Bremerhaven, 12, 413–428.
Roussel de Vauzème, D. M. (1843). Note sur l'Odontobius ceti de l'ordre des intestinaux cavitaires. Annales des Sciences Naturelles, Zoologie et biologie animale, 2(1), 326–331.
Seinhorst, J. W. (1959). A rapid method for the transfer of nematodes from fixative to anhydrous glycerin. Nematologica, 4(1), 67–69.
Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30(9), 1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Tchesunov, A. V., & Pletnikova, M. V. (1986). New data on commensal nematodes from the genus Gammarinema Kinne et Gerlach, 1953 (Chromadoria, Monhysterida). Bulletin Moskovskogo Obshchestva Ispytatelei Prirody, Otdelenie Biologiya, 91(2), 73–83.
Yoshimura, K. (1990). Two new species of the genus Monhystrium Cobb, 1920 (Monhysterida: Nematoda) from terrestrial crabs of subfamily Sesarminae (Brachyuran) in Japan. Zoological Science, 7, 755–761.
Acknowledgements
The New Caledonia Hydrobiological expedition 2016–2018 (PI: Philippe Bouchet) is a part of a cluster of expeditions under the Our Planet Reviewed/La Planète revisitée programme, implemented by the Muséum national d’Histoire naturelle; Pascale Joannot, Head of expeditions programme) in partnership with the Conservatoire d’Espaces naturels, with funding from the Gouvernement de la Nouvelle-Calédonie, Province Sud, Province Nord, Office des Postes et Télécommunications, Maison de la Nouvelle-Calédonie, and the French Ministry for the Overseas. The expedition operated under the permits issued by the Province Sud and the Province Nord, and the organisers thank, respectively, Emmanuel Coutures and Isabelle Jurquet (Province Sud) and Jean-Jérôme Cassan and Yannick Monlouis (Province Nord) for issuing of the permits. For logistics before, during, and after the field work, we thank Sébastien Faninoz and Alice Leblond. Our Planet Reviewed/La Planète revisitée is a global initiative founded in 2007 by the Muséum national d’Histoire naturelle and Pro-Natura International. The authors are grateful to Nicolas Charpin, Kaj Hnauane and Christine Pöllabauer for their invaluable assistance during 2017 and 2018 sampling trips.
Funding
Open access funding provided by Swedish Museum of Natural History. Not applicable.
Author information
Authors and Affiliations
Contributions
RW prepared species descriptions and illustrations, phylogenetic analysis. OH conceived the study and collected specimens. MA performed sequencing. All authors contributed to writing the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
All applicable institutional, national and international guidelines for the care and use of animals were followed.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Westerman, R., Ahmed, M. & Holovachov, O. Gammarinema scyllae sp. n. and Monhystrium mangrovi sp. n. (Nematoda: Monhysteridae) from land crabs from New Caledonia. Syst Parasitol 99, 83–101 (2022). https://doi.org/10.1007/s11230-021-10017-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11230-021-10017-1