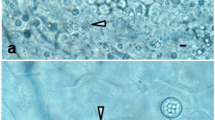
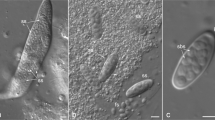
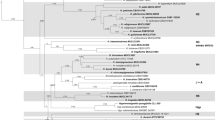
Abstract—
Numerous nucleotide sequences of microsporidia sensu lato, mainly belonging to the “Cryptomycota” (Rozellida, Rozellomycota, Rozellosporidia, treated here as synonyms), are found in metagenomes, transcriptomes, and amplicon libraries used for metabarcoding. In this study, we describe rDNA sequences of hyperparasitic metchnikovellid microsporidia found in the transcriptomes of unicellular protists belonging to Apicomplexa (Alveolata). The transcriptome of the eugregarine Polyrhabdina sp. (GenBank SRX6640468) contains the cDNA of Metchnikovella incurvata, the transcriptome of the archigregarine Selenidium pygospionis (GenBank SRX6640459) contains the cDNA of Metchnikovella dogieli, and in the transcriptome of the blastogregarine Siedleckia cf. nematoides (GenBank SRX6640464) we find cDNAs originating from a yet undescribed species representing a novel metchnikovellid family. We have modeled the secondary structure of the “ITS2” region of identified and unidentified metchnikovellids taking into account the covariant nucleotide substitutions. Based on the predicted secondary structure of rRNA, mapping of reads from cDNA libraries, and the absence of the endoribonuclease Las1 (PF04031), we conclude that there is no ITS2 processing in metchnikovellids, and the mature “5.8S”- and “28S”-like (LSU) rRNA are covalently fused, similarly to the LSU rRNA in the other microsporidia sensu stricto. We discuss several previously proposed (Chytridiopsis typographi, BAQA065) and new candidates for the sister group of microsporidia sensu stricto, and compare the reduced rRNA genes of microsporidia and the lengthened rRNA genes with group I introns of parasitic and lichen fungi in the context of neutral and adaptive evolutionary processes.












Similar content being viewed by others
REFERENCES
Aleoshin, V.V., Mikhailov, K.V., and Karpov, S.A., On the origin and early evolution of fungi and microsporidia, in Sbornik nauchno-populyarnykh statei i fotomaterialov—pobeditelei konkursa RFFI 2015 goda (Collection of Popular Scientific Papers and Photographs of Competition Winners of the Russian Foundation of Basic Research in 2015), Moscow: Molnet, 2015, no. 18, pp. 215–223.
Aleshin, V.V., Konstantinova, A.V., Mikhailov, K.V., Nikitin, M.A., and Petrov, N.B., Do we need many genes for phylogenetic inference? Biochemistry (Moscow), 2007, vol. 72, no. 12, pp. 1313–1323.
Alkemar, G. and Nygård, O., A possible tertiary rRNA interaction between expansion segments ES3 and ES6 in eukaryotic 40S ribosomal subunits, RNA, 2003, vol. 9, no. 1, pp. 20–24.
Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., et al., Gapped BLAST and PSI-BLAST: a new generation of protein database search programs, Nucleic Acids Res., 1997, vol. 25, no. 17, pp. 3389–3402.
Antony, C.P., Kumaresan, D., Hunger, S., Drake, H.L., Murrell, J.C., and Shouche, Y.S., Microbiology of Lonar Lake and other soda lakes, ISME J., 2013, vol. 7, no. 3, pp. 468–476.
Arroyo, A.S., López-Escardó, D., Kim, E., Ruiz-Trillo, I., and Najle, S.R., Novel diversity of deeply branching Holomycota and unicellular holozoans revealed by metabarcoding in Middle Paraná River, Argentina, Front. Ecol. Evol., 2018, vol. 6, art. ID 99.
Barandun, J., Hunziker, M., Vossbrinck, C.R., and Klinge, S., Evolutionary compaction and adaptation visualized by the structure of the dormant microsporidian ribosome, Nat. Microbiol., 2019, vol. 4, no. 11. pp. 1798–1804.
Bass, D., Czech, L., Williams, B.A.P., Berney, C., Dunthorn, M., et al., Clarifying the relationships between Microsporidia and Cryptomycota, J. Eukaryotic Microbiol., 2018, vol. 65, no. 6, pp. 773–782.
Benson, D.A., Cavanaugh, M., Clark, K., Karsch-Mizrachi, I., Lipman, D.J., et al., GenBank, Nucleic Acids Res., 2013, vol. 41, no. 1, pp. D36–D42.
Berney, C., Fahrni, J., and Pawlowski, J., How many novel eukaryotic ‘kingdoms’? Pitfalls and limitations of environmental DNA surveys, BMC Biol., 2004, vol. 2, art. ID 13.
Bernhart, S.H., Hofacker, I.L., Will, S., Gruber, A.R., and Stadler, P.F., RNAalifold: improved consensus structure prediction for RNA alignments, BMC Bioinf., 2008, vol. 9, art. ID 474.
Bhattacharya, D., Friedl, T., and Damberger, S., Nuclear-encoded rDNA group I introns: origin and phylogenetic relationships of insertion site lineages in the green algae, Mol. Biol. Evol., 1996, vol. 13, no. 7, pp. 978–989.
Bhattacharya, D., Friedl, T., and Helms, G., Vertical evolution and intragenic spread of lichen-fungal group I introns, J. Mol. Evol., 2002, vol. 55, no. 1, pp. 74–84.
Borner, J. and Burmester, T., Parasite infection of public databases: a data mining approach to identify apicomplexan contaminations in animal genome and transcriptome assemblies, BMC Genomics, 2017, vol. 18, no. 1, art. ID 100.
Burki, F., Corradi, N., Sierra, R., Pawlowski, J., Meyer, G.R., et al., Phylogenomics of the intracellular parasite Mikrocytos mackini reveals evidence for a mitosome in Rhizaria, Curr. Biol., 2013, vol. 23, no. 16, pp. 1541–1547.
Burki, F., Kaplan, M., Tikhonenkov, D.V., Zlatogursky, V., Minh, B.Q., et al., Untangling the early diversification of eukaryotes: a phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista, Proc. R. Soc. B, 2016, vol. 283, no. 1823, art. ID 20152802.
Carnegie, R.B., Meyer, G.R., Blackbourn, J., Cochennec-Laureau, N., Berthe, F.C., and Bower, S.M., Molecular detection of the oyster parasite Mikrocytos mackini, and a preliminary phylogenetic analysis, Dis. Aquat. Org., 2003, vol. 54, no. 3, pp. 219–227.
Cavalier-Smith, T. and Chao, E.E., Phylogeny and classification of phylum Cercozoa (Protozoa), Protist, 2003, vol. 154, nos. 3–4, pp. 341–358.
Chaker-Margot M., Assembly of the small ribosomal subunit in yeast: mechanism and regulation, RNA, 2018, vol. 24, no. 7, pp. 881–891.
Chalwatzis, N., Baur, A., Stetzer, E., Kinzelbach, R., and Zimmermann, F.K., Strongly expanded 18S rRNA genes correlated with a peculiar morphology in the insect order of Strepsiptera, Zoology, 1995, vol. 98, no. 2, pp. 115–126.
Chorev, M. and Carmel, L., The function of introns, Front. Genet., 2012, vol. 3, art. ID 55.
Chouari, R., Leonard, M., Bouali, M., Guermazi, S., Rahli, N., et al., Eukaryotic molecular diversity at different steps of the wastewater treatment plant process reveals more phylogenetic novel lineages, World J. Microbiol. Biotechnol., 2017, vol. 33, no. 3, art. ID 44.
Christaki, U., Genitsaris, S., Monchy, S., Li, L.L., Rachik, S., et al., Parasitic eukaryotes in a meso-eutrophic coastal system with marked Phaeocystis globosa blooms, Front. Mar. Sci., 2017, vol. 4, art. ID 416.
Chupov, V.S., Form of the lateral phylogenetic branch in plants, Usp. Sovrem. Biol., 2002, vol. 122, no. 3, pp. 227–238.
Coleman, A.W., Pan-eukaryote ITS2 homologies revealed by RNA secondary structure, Nucleic Acids Res., 2007, vol. 35, no. 10, pp. 3322–3329.
Coleman, A.W., Nuclear rRNA transcript processing versus internal transcribed spacer secondary structure, Trends Genet., 2015, vol. 31, no. 3, pp. 157–163.
Corsaro, D., Walochnik, J., Venditti, D., Müller, K.-D., Hauröder, B., and Michel, R., Rediscovery of Nucleophaga amoebae, a novel member of the Rozellomycota, Parasitol. Res., 2014a, vol. 113, no. 12, pp. 4491–4498.
Corsaro, D., Walochnik, J., Venditti, D., Steinmann, J., Müller, K.D., and Michel, R., Microsporidia-like parasites of amoebae belong to the early fungal lineage Rozellomycota, Parasitol. Res., 2014b, vol. 113, no. 5, pp. 1909–1918.
Corsaro, D., Michel, R., Walochnik, J., Venditti, D., Müller, K.D., et al., Molecular identification of Nucleophaga terricolae sp. nov. (Rozellomycota), and new insights on the origin of the microsporidia, Parasitol. Res., 2016, vol. 115, no. 8, pp. 3003–3011.
Corsaro, D., Wylezich, C., Venditti, D., Michel, R., Walochnik, J., and Wegensteiner, R., Filling gaps in the microsporidian tree: rDNA phylogeny of Chytridiopsis typographi (Microsporidia: Chytridiopsida), Parasitol. Res., 2019, vol. 118, no. 1, pp. 169–180.
Corsaro, D., Walochnik, J., Venditti, D., Hauröder, B., and Michel, R., Solving an old enigma: Morellospora saccamoebae gen. nov., sp. nov. (Rozellomycota), a Sphaerita-like parasite of free-living amoebae, Parasitol. Res., 2020, vol. 119, no. 3, pp. 925–934.
Cotto, I., Dai, Z., Huo, L., Anderson, C.L., Vilardi, K.J., et al., Long solids retention times and attached growth phase favor prevalence of comammox bacteria in nitrogen removal systems, Water Res., 2020, vol. 169, art. ID 115268.
Dawson, S.C. and Pace, N.R., Novel kingdom-level eukaryotic diversity in anoxic environments, Proc. Natl. Acad. Sci. U.S.A., 2002, vol. 99, no. 12, pp. 8324–8329.
Delalibera, I., Jr., Hajek, A.E., and Humber, R.A., Neozygites tanajoae sp. nov., a pathogen of the cassava green mite, Mycologia, 2004, vol. 96, no. 5, pp. 1002–1009.
DePriest, P.T. and Been, M.D., Numerous group I introns with variable distributions in the ribosomal DNA of a lichen fungus, J. Mol. Biol., 1992, vol. 228, no. 2, pp. 315–321.
De Rijk, P., Gatehouse, H.S., and De Wachter, R., The secondary structure of Nosema apis large subunit ribosomal RNA, Biochim. Biophys. Acta, Gene Struct. Expression, 1998, vol. 1442, nos. 1–2, pp. 326–328.
De Rijk, P., Wuyts, J., and De Wachter, R., RnaViz 2: an improved representation of RNA secondary structure, Bioinformatics, 2003, vol. 19, no. 2, pp. 299–300.
Edgar, R.C., MUSCLE: multiple sequence alignment with high accuracy and high throughput, Nucleic Acids Res., 2004, vol. 32, no. 5, pp. 1792–1797.
Eichorst, S.A. and Kuske, C.R., Identification of cellulose-responsive bacterial and fungal communities in geographically and edaphically different soils by using stable isotope probing, Appl. Environ. Microbiol., 2012, vol. 78, no. 7, pp. 2316–2327.
Engel, C., Sainsbury, S., Cheung A.C., Kostrewa, D., and Cramer, P., RNA polymerase I structure and transcription regulation, Nature, 2013, vol. 502, no. 7473, pp. 650–655.
Felsenstein, J., Cases in which parsimony or compatibility methods will be positively misleading, Syst. Biol., 1978, vol. 27, no. 4, pp. 401–410.
Fernández-Tornero, C., RNA polymerase I activation and hibernation: unique mechanisms for unique genes, Transcription, 2018, vol. 9, no. 4, pp. 248–254.
Filée, J., Tetart, F., Suttle, C.A., and Krisch, H.M., Marine T4-type bacteriophages, a ubiquitous component of the dark matter of the biosphere, Proc. Natl. Acad. Sci. U.S.A., 2005, vol. 102, no. 35, pp. 12471–12476.
Fitch, D.H., Bugaj-Gaweda, B., and Emmons, S.W., 18S ribosomal RNA gene phylogeny for some Rhabditidae related to Caenorhabditis, Mol. Biol. Evol., 1995, vol. 12, no. 2, pp. 346–358.
Förster, J., Famili, I., Fu, P., Palsson, B.Ø., and Nielsen, J., Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network, Genome Res., 2003, vol. 13, no. 2, pp. 244–253.
Freimoser, F.M., Cultivation, sporulation and phylogenetic analysis of Neozygites parvispora and Entomophthora thripidum, two fungal pathogens of thrips, PhD Thesis, Zürich: ETH Zürich, 2000, pp. 29–43. https://www.research-collection.ethz.ch/handle/20.5-00.11850/144847.
Galindo, L.J., Torruella, G., Moreira, D., Timpano, H., Paskerova, G., et al., Evolutionary genomics of Metchnikovella incurvata (Metchnikovellidae): an early branching microsporidium, Genome Biol. Evol., 2018, vol. 10, no. 10, pp. 2736–2748.
Gargas, A., DePriest, P.T., and Taylor, J.W., Positions of multiple insertions in SSU rDNA of lichen-forming fungi, Mol. Biol. Evol., 1995, vol. 12, no. 2, pp. 208–218.
Gawryluk, R.M.R., Tikhonenkov, D.V., Hehenberger, E., Husnik, F., Mylnikov, A.P., and Keeling, P.J., Non-photosynthetic predators are sister to red algae, Nature, 2019, vol. 572, no. 7768, pp. 240–243.
Gleason, F.H., Lilje, O., Marano, A.V., Sime-Ngando, T., Sullivan, B.K., et al., Ecological functions of zoosporic hyperparasites, Front. Microbiol., 2014, vol. 5, art. ID 244.
Gromov, B.V., Algae parasites from the Tsenovskii “monad” group of genera Aphelidium, Amoeboaphelidium, and Pseudaphelidium as members of a new class, Zool. Zh., 2000, vol. 79, no. 5, pp. 517–525.
Grossart, H.-P., Wurzbacher, C., James, T.Y., and Kagami, M., Discovery of dark matter fungi in aquatic ecosystems demands a reappraisal of the phylogeny and ecology of zoosporic fungi, Fungal Ecol., 2016, vol. 19, no. 1, pp. 28–38.
Gruber, A.R., Bernhart, S.H., and Lorenz, R., The ViennaRNA web services, Methods Mol. Biol., 2015, vol. 1269, pp. 307–326.
Guillou, L., Viprey, M., Chambouvet, A., Welsh, R.M., Kirkham, A.R., et al., Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata), Environ. Microbiol., 2008, vol. 10, no. 12, pp. 3349–3365.
Haag, K.L., James, T.Y., Pombert, J.F., Larsson, R., Schaer, T.M., et al., Evolution of a morphological novelty occurred before genome compaction in a lineage of extreme parasites, Proc. Natl. Acad. Sci. U.S.A., 2014, vol. 111, no. 43, pp. 15480–15485.
Hannen van, E.J., Mooij, W., van Agterveld, M.P., Gons, H.J., and Laanbroek, H.J., Detritus-dependent development of the microbial community in an experimental system: qualitative analysis by denaturing gradient gel electrophoresis, Appl. Environ. Microbiol., 1999, vol. 65, no. 6, pp. 2478–2484.
Hartmann, M., Lee, S., Hallam, S.J., and Mohn, W.W., Bacterial, archaeal and eukaryal community structures throughout soil horizons of harvested and naturally disturbed forest stands, Environ. Microbiol., 2009, vol. 11, no. 12, pp. 3045–3062.
Hasegawa, H., Satô, M., Maeda, K., and Murayama, Y., Description of Riouxgolvania kapapkamui sp. n. (Nematoda: Muspiceoidea: Muspiceidae), a peculiar intradermal parasite of bats in Hokkaido, Japan, J. Parasitol., 2012, vol. 98, no. 5, pp. 995–1000.
Hendy, M.D. and Penny, D., A framework for the quantitative study of evolutionary trees, Syst. Zool., 1989, vol. 38, no. 4, pp. 277–290.
Hennig, W., Phylogenetic Systematics, Urbana: Univ. of Illinois Press, 1966.
Huss, V.A.R. and Bauer, C., A highly divergent 18S rRNA sequence identified by environmental PCR from an extremely acidic mining lake (pH 2.3) in Lusatia (Germany), 2011, no. FN546176.1. https://www.ncbi.nlm.nih. gov/nuccore/345107473.
Ishida, S., Nozaki, D., Grossart, H.P., and Kagami, M., Novel basal, fungal lineages from freshwater phytoplankton and lake samples, Environ. Microbiol. Rep., 2015, vol. 7, no. 3, pp. 435–441.
Issi, I.V., Development of microsporidiology in Russia, Vestn. Zashch. Rast., 2020, vol. 103, no. 3, pp. 161–176.
Issi, I.V. and Voronin, V.N., Type Microsporidia Balbiani, 1882, in Protisty: rukovodstvo po zoologii (Protists: A Guide on Zoology), Alimov, A.F., Ed., St. Petersburg: Nauka, 2007, part 2, pp. 994–1045.
James, T.Y., Kauff, F., Schoch, C.L., Matheny, P.B., Hofstetter, V., et al., Reconstructing the early evolution of Fungi using a six-gene phylogeny, Nature, 2006, vol. 443, no. 7113, pp. 818–822.
Jamy, M., Foster, R., Barbera, P., Czech, L., Kozlov, A., et al., Long-read metabarcoding of the eukaryotic rDNA operon to phylogenetically and taxonomically resolve environmental diversity, Mol. Ecol. Resour., 2019, vol. 20, no. 2, pp. 429–443.
Janouškovec, J., Tikhonenkov, D.V., Burki, F., Howe, A.T., Rohwer, F.L., et al., A new lineage of eukaryotes illuminates early mitochondrial genome reduction, Curr. Biol., 2017, vol. 27, no. 23, pp. 3717–3724.
Janouškovec, J., Paskerova, G.G., Miroliubova, T.S., Mikhailov, K.V., Birley, T., et al., Apicomplexan-like parasites are polyphyletic and widely but selectively dependent on cryptic plastid organelles, eLife, 2019, vol. 8, art. ID e49662.
Jones, M.D.M., Forn, I., Gadelha, C., Egan, M.J., Bass, D., et al., Discovery of novel intermediate forms redefines the fungal tree of life, Nature, 2011a, vol. 474, no. 7350, pp. 200–203.
Jones, M.D.M., Richards, T.A., Hawksworth, D.J., and Bass, D., Validation of the phylum name Cryptomycota phyl. nov. with notes on its recognition, IMA Fungus, 2011b, vol. 2, no. 2, pp. 173–175.
Joseph, N., Krauskopf, E., Vera, M.I., and Michot, B., Ribosomal internal transcribed spacer 2 (ITS2) exhibits a common core of secondary structure in vertebrates and yeast, Nucleic Acids Res., 1999, vol. 27, no. 23, pp. 4533–4540.
Karpov, S.A. and Paskerova, G.G., The aphelids, intracellular parasitoids of algae, consume the host cytoplasm “from the inside,” Protistology, 2020, vol. 14, no. 4, pp. 258–263.
Karpov, S.A., Mikhailov, K.V., Mirzaeva, G.S., Mirabdullaev, I.M., Mamkaeva, K.A., et al., Obligately phagotrophic aphelids turned out to branch with the earliest-diverging fungi, Protist, 2013, vol. 164, no. 2, pp. 195–205.
Karpov, S.A., Mamanazarova, K.S., Popova, O.V., Aleoshin, V.V., James, T.Y., et al., Monoblepharidomycetes diversity includes new parasitic and saprotrophic species with highly intronized rDNA, Fungal Biol., 2017, vol. 121, no. 8, pp. 729–741.
Karpov, S.A., Moreira, D., Mamkaeva, M.A., Popova, O.V., Aleoshin, V.V., and López-García, P., New member of Gromochytriales (Chytridiomycetes)–Apiochytrium granulosporum nov. gen. et sp., J. Eukaryotic Microbiol., 2019, vol. 66, no. 4, pp. 582–591.
Katiyar, S.K., Visvesvara, G.S., and Edlind, T.D., Comparisons of ribosomal RNA sequences from amitochondrial protozoa: implications for processing, mRNA binding and paromomycin susceptibility, Gene, 1995, vol. 152, no. 1, pp. 27–33.
Kim, E., Sprung, B., Duhamel, S., Filardi, C., and Kyoon Shin, M., Oligotrophic lagoons of the South Pacific Ocean are home to a surprising number of novel eukaryotic microorganisms, Environ. Microbiol., 2016, vol. 18, no. 12, pp. 4549–4563.
Kimura, M., The Neutral Theory of Molecular Evolution, Cambridge: Cambridge Univ. Press, 1983.
Kumar, S., Stecher, G., and Tamura, K., MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets, Mol. Biol. Evol., 2016, vol. 33, no. 7, pp. 1870–1874.
Langmead, B. and Salzberg, S.L., Fast gapped-read alignment with Bowtie 2, Nat. Methods, 2012, vol. 9, no. 4, pp. 357–359.
Lara, E., Moreira, D., and López-García, P., The environmental clade LKM11 and Rozella form the deepest branching clade of fungi, Protist, 2010, vol. 161, no. 1, pp. 116–121.
Larsson, J.I.R., Description of Chytridiopsis trichopterae n. sp. (Microspora, Chytridiopsidae), a microsporidian parasite of the caddis fly Polycentropus flavomaculatus (Trichoptera, Polycentropodidae), with comments on relationships between the families Chytridiopsidae and Metchnikovellidae, J. Eukaryotic Microbiol., 1993, vol. 40, no. 1, pp. 37–48.
Larsson, J.I.R., The hyperparasitic microsporidium Amphiacantha longa Caullery et Mesnil, 1914 (Microspora: Metchnikovellidae)—description of the cytology, redescription of the species, emended diagnosis of the genus Amphiacantha and establishment of the new family Amphiacanthidae, Folia Parasitol., 2000, vol. 47, no. 4, pp. 241–256.
Larsson, J.I.R., The primitive Microsporidia, in Microsporidia: Pathogens of Opportunity, Weiss, L.M. and Becnel, J.J., Eds., Oxford: Wiley-Blackwell, 2014, pp. 605–634.
Lazarus, K.L. and James, T.Y., Surveying the biodiversity of the Cryptomycota using a targeted PCR approach, Fungal Ecol., 2015, vol. 14, no. 1, pp. 62–70.
Lee, J.C. and Gutell, R.R., A comparison of the crystal structures of the eukaryotic and bacterial SSU ribosomal RNAs reveals common structural features in the hypervariable regions, PLoS One, 2012, vol. 7, no. 5, art. ID e38203.
Lefèvre, E., Bardot, C., Noël, C., Carrias, J.F., Viscogliosi, E., et al., Unveiling fungal zooflagellates as members of freshwater picoeukaryotes: evidence from a molecular diversity study in a deep meromictic lake, Environ. Microbiol., 2007, vol. 9, no. 1, pp. 61–71.
Lefèvre, E., Roussel, B., Amblard, C., and Sime-Ngando, T., The molecular diversity of freshwater picoeukaryotes reveals high occurrence of putative parasitoids in the plankton, PLoS One, 2008, vol. 3, no. 6, art. ID e2324.
Leipe, D.D., Gunderson, J.H., Nerad, T.A., and Sogin, M.L., Small subunit ribosomal RNA+ of Hexamita inflata and the quest for the first branch in the eukaryotic tree, Mol. Biochem. Parasitol., 1993, vol. 59, no. 1, pp. 41–48.
Lepère, C., Boucher, D., Jardillier, L., Domaizon, I., and Debroas, D., Succession and regulation factors of small eukaryote community composition in a lacustrine ecosystem (Lake Pavin), Appl. Environ. Microbiol., 2006, vol. 72, no. 4, pp. 2971–2981.
Lepère, C., Domaizon, I., and Debroas, D., Unexpected importance of potential parasites in the composition of the freshwater small-eukaryote community, Appl. Environ. Microbiol., 2008, vol. 74, no. 10, pp. 2940–2949.
Letcher, P.M. and Powell, M.J., A taxonomic summary and revision of Rozella (Cryptomycota), IMA Fungus, 2018, vol. 9, pp. 383–399.
Letcher, P.M., Lopez, S., Schmieder, R., Lee, P.A., Behnke, C., et al., Characterization of Amoeboaphelidium protococcarum, an algal parasite new to the cryptomycota isolated from an outdoor algal pond used for the production of biofuel, PLoS One, 2013, vol. 8, no. 2, art. ID e56232.
Lipson, D.A., Kuske, C.R., Gallegos-Graves, L.V., and Oechel, W.C., Elevated atmospheric CO2 stimulates soil fungal diversity through increased fine root production in a semiarid shrubland ecosystem, Global Change Biol., 2014, vol. 20, no. 8, pp. 2555–2565.
López-García, P., Rodriguez-Valera, F., Pedrós-Alió, C., and Moreira, D., Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton, Nature, 2001, vol. 409, no. 6820, pp. 603–607.
Mahé, F., de Vargas, C., Bass, D., Czech, L., Stamatakis, A., et al., Parasites dominate hyperdiverse soil protist communities in Neotropical rainforests, Nat. Ecol. Evol., 2017, vol. 1, no. 4, art. ID 91.
Mamkaev, Yu.V., Comparison of morphological differences in the lower and higher groups of the same phylogenetic branch, Zh. Obshch. Biol., 1968, vol. 29, no. 1, pp. 48–56.
Marcy, Y., Ouverney, C., Bik, E.M., Lösekann, T., Ivanova, N., et al., Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth, Proc. Natl. Acad. Sci. U.S.A., 2007, vol. 104, no. 29, pp. 11889–11894.
Matsubayashi, M., Shimada, Y., Li, Y.Y., Harada, H., and Kubota, K., Phylogenetic diversity and in situ detection of eukaryotes in anaerobic sludge digesters, PLoS One, 2017, vol. 12, no. 3, art. ID e0172888.
McGuire, K.L., Allison, S.D., Fierer, N., and Treseder, K., Ectomycorrhizal-dominated boreal and tropical forests have distinct fungal communities, but analogous spatial patterns across soil horizons, PLoS One, 2013, vol. 8, no. 7, art. ID e68278.
Melnikov, S.V., Manakongtreecheep, K., Rivera, K.D., Makarenko, A., Pappin, D.J., and Söll, D., Muller’s ratchet and ribosome degeneration in the obligate intracellular parasites Microsporidia, Int. J. Mol. Sci., 2018a, vol. 19, no. 12, art. ID 4125.
Melnikov, S.V., Rivera, K.D., Ostapenko, D., Makarenko, A., Sanscrainte, N.D., et al., Error-prone protein synthesis in parasites with the smallest eukaryotic genome, Proc. Natl. Acad. Sci. U.S.A., 2018b, vol. 115, no. 27, pp. E6245–E6253.
Mikhailov, K.V., Simdyanov, T.G., and Aleoshin, V.V., Genomic survey of a hyperparasitic microsporidian Amphiamblys sp. (Metchnikovellidae), Genome Biol. Evol., 2017, vol. 9, no. 3, pp. 454–467.
Mitchell, A.L., Scheremetjew, M., Denise, H., Potter, S., Tarkowska, A., et al., EBI Metagenomics in 2017: enriching the analysis of microbial communities, from sequence reads to assemblies, Nucleic Acids Res., 2018, vol. 46, no. 1, pp. D726–D735.
Monchy, S., Sanciu, G., Jobard, M., Rasconi, S., Gerphagnon, M., et al., Exploring and quantifying fungal diversity in freshwater lake ecosystems using rDNA cloning/sequencing and SSU tag pyrosequencing, Environ. Microbiol., 2011, vol. 13, no. 6, pp. 1433–1453.
Moon-van der Staay, S.Y., De Wachter, R., and Vaulot, D., Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity, Nature, 2001, vol. 409, no. 6820, pp. 607–610.
Moss, T. and Stefanovsky, V.Y., At the center of eukaryotic life, Cell, 2002, vol. 109, no. 5, pp. 545–548.
Mueller, R.C., Moya-Balasch, M., and Kuske, C.R., Contrasting soil fungal community responses to experimental nitrogen addition using the large subunit rRNA taxonomic marker and cellobiohydrolase I functional marker, Mol. Ecol., 2014, vol. 23, no. 17, pp. 4406–4417.
Najmi, S.M. and Schneider, D.A., Quorum sensing regulates rRNA synthesis in Saccharomyces cerevisiae, Gene, 2021, vol. 776, art. ID 145442.
Nakai, R., Abe, T., Baba, T., Imura, S., Kagoshima, H., et al., Eukaryotic phylotypes in aquatic moss pillars inhabiting a freshwater lake in East Antarctica, based on 18S rRNA gene analysis, Polar Biol., 2012, vol. 35, no. 10, pp. 1495–1504.
Nassonova, E.S., Bondarenko, N.I., Paskerova, G.G., Kovacikova, M., Frolova, E.V., and Smirnov, A.V., Evolutionary relationships of Metchnikovella dogieli Paskerova et al., 2016 (Microsporidia: Metchnikovellidae) revealed by multigene phylogenetic analysis, Parasitol. Res., 2021, vol. 120, no. 2, pp. 525–534.
Noack Watt, K.E., Achilleos, A., Neben, C.L., Merrill, A.E., and Trainor, P.A., The roles of RNA polymerase I and III subunits Polr1c and Polr1d in craniofacial development and in zebrafish models of Treacher Collins syndrome, PLoS Genet., 2016, vol. 12, no. 7, art. ID e1006187.
Page, K.A. and Flannery, M.K., Microbial epiphytes of deep-water moss in Crater Lake, Oregon, Northwest Sci., 2018, vol. 92, no. 4, pp. 240–250.
Paskerova, G.G., Frolova, E.V., Kováčiková, M., Panfilkina, T.S., Mesentsev, E.S., et al., Metchnikovella dogieli sp. n. (Microsporidia: Metchnikovellida), a parasite of archigregarines Selenidium sp. from polychaetes Pygospio elegans, Protistology, 2016a, vol. 10, no. 4, pp. 148–157.
Paskerova, G.G., Miroliubova, T.S., Diakin, A., Kováčiková, M., Valigurová, A., et al., Fine structure and molecular phylogenetic position of two marine gregarines, Selenidium pygospionis sp. n. and S. pherusae sp. n., with notes on the phylogeny of Archigregarinida (Apicomplexa), Protist, 2016b, vol. 169, no. 6, pp. 826–852.
Pawlowski, J., Bolivar, I., Guiard-Maffia, J., and Gouy, M., Phylogenetic position of foraminifera inferred from LSU rRNA gene sequences, Mol. Biol. Evol., 1994, vol. 11, no. 6, pp. 929–938.
Pawlowski, J., Montoya-Burgos, J.I., Fahrni, J.F., Wüest, J., and Zaninetti, L., Origin of the Mesozoa inferred from 18S rRNA gene sequences, Mol. Biol. Evol., 1996, vol. 13, no. 8, pp. 1128–1132.
Peer van de, Y., Ben Ali, A., and Meyer, A., Microsporidia: accumulating molecular evidence that a group of amitochodriate and suspectedly primitive eukaryotes are just curious fungi, Gene, 2000, vol. 246, nos. 1–2, pp. 1–8.
Petrov, A.S., Bernier, C.R., Gulen, B., Waterbury, C.C., Hershkovits, E., et al., Secondary structures of rRNAs from all three domains of life, PLoS One, 2014, vol. 9, no. 2, art. ID e88222.
Peyretaillade, E., Biderre, C., Peyret, P., Duffieux, F., Méténier, G., et al., Microsporidian Encephalitozoon cuniculi, a unicellular eukaryote with an unusual chromosomal dispersion of ribosomal genes and a LSU rRNA reduced to the universal core, Nucleic Acids Res., 1998, vol. 26, no. 15, pp. 3513–3520.
Quinlan, A.R. and Hall, I.M., BEDTools: a flexible suite of utilities for comparing genomic features, Bioinformatics, 2010, vol. 26, no. 6, pp. 841–842.
Rambaut, A., FigTree v1.3.1, Edinburgh Institute of Evolutionary Biology, University of Edinburgh, 2010. http://tree.bio.ed.ac.uk/software/figtree/.
Rogers, S.O., Integrated evolution of ribosomal RNAs, introns, and intron nurseries, Genetica, 2019, vol. 147, no. 2, pp. 103–119.
Rojas-Jimenez, K., Wurzbacher, C., Bourne, E.C., Chiuchiolo, A., Priscu, J.C., and Grossart, H.P., Early diverging lineages within Cryptomycota and Chytridiomycota dominate the fungal communities in ice-covered lakes of the McMurdo Dry Valleys, Antarctica, Sci. Rep., 2017, vol. 7, no. 1, art. ID 15348.
Rojas-Jimenez, K., Rieck, A., Wurzbacher, C., Jürgens, K., Labrenz, M., and Grossart, H.P., A salinity threshold separating fungal communities in the Baltic Sea, Front. Microbiol., 2019, vol. 10, art. ID 680.
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., et al., MrBayes 3.2: Efficient Bayesian phylogenetic inference and model selection across a large model space, Syst. Biol., 2012, vol. 61, no. 3, pp. 539–542.
Rotari, Y.M., Paskerova, G.G., and Sokolova, Y.Y., Diversity of metchnikovellids (Metchnikovellidae, Rudimicrosporea), hyperparasites of bristle worms (Annelida, Polychaeta) from the White Sea, Protistology, 2015, vol. 9, no. 1, pp. 50–59.
Rueckert, S., Simdyanov, T.G., Aleoshin, V.V., and Leander, B.S., Identification of a divergent environmental DNA sequence clade using the phylogeny of gregarine parasites (Apicomplexa) from crustacean hosts, PLoS One, 2011, vol. 6, no. 3, art. ID e18163.
Rueckert, S., Wakeman, K.C., and Leander, B.S., Discovery of a diverse clade of gregarine apicomplexans (Apicomplexa: Eugregarinorida) from Pacific eunicid and onuphid polychaetes, including descriptions of Paralecudina n. gen., Trichotokara japonica n. sp., and T. eunicae n. sp., J. Eukaryotic Microbiol., 2013, vol. 60, no. 2, pp. 121–136.
Russell, J. and Zomerdijk, J.C.B.M., RNA-polymerase-I-directed rDNA transcription, life and works, Trends Biochem. Sci., 2005, vol. 30, no. 2, pp. 87–96.
Sanchez, L.R.S. and Cao, E.P., Metagenomic analysis reveals the presence of heavy metal response genes from cyanobacteria thriving in Balatoc Mines, Benguet Province, Philippines, Philipp. J. Sci., 2019, vol. 148, suppl. 1, pp. 71–82.
Schultz, J., Maisel, S., Gerlach, D., Műller, T., and Wolf, M., A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the eukaryote, RNA, 2005, vol. 11, no. 4, pp. 361–364.
Seenivasan, R., Sausen, N., Medlin, L.K., and Melkonian, M., Picomonas judraskeda gen. et sp. nov.: the first identified member of the Picozoa phylum nov., a widespread group of picoeukaryotes, formerly known as ‘picobiliphytes,’ PLoS One, 2013, vol. 8, no. 3, art. ID e59565.
Semenov, M.V., Metabarcoding and metagenomics in soil ecology research: achievements, challenges, and prospects, Biol. Bull. Rev., 2021, vol. 11, no. 1, pp. 40–53.
Sharrar, A.M., Crits-Christoph, A., Méheust, R., Diamond, S., Starr, E.P., and Banfield, J.F., Bacterial secondary metabolite biosynthetic potential in soil varies with phylum, depth, and vegetation type, mBio, 2020, vol. 11, no. 3, art. ID e00416-20.
Smith, M.E., Douhan, G.W., and Rizzo, D.M., Ectomycorrhizal community structure in a xeric Quercus woodland as inferred from rDNA sequence analysis of pooled EM roots and sporocarps, New Phytol., 2007, vol. 174, no. 4, pp. 847–863.
Smothers, J.F., von Dohlen, C.D., Smith, L.H., Jr., and Spall, R.D., Molecular evidence that the myxozoan protists are metazoans, Science, 1994, vol. 265, no. 5179, pp. 1719–1721.
Sokolova, Y.Y., Paskerova, G.G., Rotari, Y.M., Nassonova, E.S., and Smirnov, A.V., Fine structure of Metchnikovella incurvata Caullery and Mesnil 1914 (Microsporidia), a hyperparasite of gregarines Polyrhabdina sp. from the polychaete Pygospio elegans, Parasitology, 2013, vol. 140, no. 7, pp. 855–867.
Sprague, V., Classification and phylogeny of the microsporidia, in Comparative Pathobiology, Vol. 2: Systematics of the Microsporidia, Bulla, L.A. and Cheng, T.C., Eds., New York: Plenum, 1977, pp. 1–30.
Stentiford, G.D., Ramilo, A., Abollo, E., Kerr, R., Bateman, K.S., et al., Hyperspora aquatica n. gen., n. sp. (Microsporidia), hyperparasitic in Marteilia cochillia (Paramyxida), is closely related to crustacean-infecting microspordian taxa, Parasitology, 2017, vol. 144, no. 2, pp. 186–199.
Stevenson, B.S. and Schmidt, T.M., Life history implications of rRNA gene copy number in Escherichia coli, Appl. Environ. Microbiol., 2004, vol. 70, no. 11, pp. 6670–6677.
Strassert, J.F.H., Jamy, M., Mylnikov, A.P., Tikhonenkov, D.V., and Burki, F., New phylogenomic analysis of the enigmatic phylum Telonemia further resolves the Eukaryote Tree of Life, Mol. Biol. Evol., 2019, vol. 36, no. 4, art. ID 757765.
Tavaré, S., Some probabilistic and statistical problems in the analysis of DNA sequences, Lectures Math. Life Sci., 1986, vol. 17, pp. 57–86.
Taylor, D.L., Herriott, I.C., Long, J., and O’Neill, K., TOPO TA is A-OK: a test of phylogenetic bias in fungal environmental clone library construction, Environ. Microbiol., 2007, vol. 9, no. 5, pp. 1329–1334.
Taylor, D.L., Booth, M.G., McFarland, J.W., Herriott, I.C., Lennon, N.J., et al., Increasing ecological inference from high throughput sequencing of fungi in the environment through a tagging approach, Mol. Ecol. Resour., 2008, vol. 8, no. 4, pp. 742–752.
Taylor, D.L., Hollingsworth, T.N., McFarland, J.W., Lennon, N.J., Nusbaum, C., and Ruess, R.W., A first comprehensive census of fungi in soil reveals both hyperdiversity and fine-scale niche partitioning, Ecol. Monogr., 2014, vol. 84, no. 1, pp. 3–20.
Tedersoo, L., Bahram, M., Puusepp, R., Nilsson, R.H., and James, T.Y., Novel soil-inhabiting clades fill gaps in the fungal tree of life, Microbiome, 2017, vol. 5, no. 1, art. ID 42.
Tedersoo, L., Sánchez-Ramírez, S., Kõljalg, U., Bahram, M., Döring, M., et al., High-level classification of the fungi and a tool for evolutionary ecological analyses, Fungal Diversity, 2018, vol. 90, no. 1, pp. 135–159.
Thornton, C.N., Tanner, W.D., VanDerslice, J.A., and Brazelton, W.J., Localized effect of treated wastewater effluent on the resistome of an urban watershed, Gigascience, 2020, vol. 9, no. 11, art. ID giaa125.
Tikhonenkov, D.V., Mikhailov, K.V., Hehenberger, E., Karpov, S.A., Prokina, K.I., et al., New lineage of microbial predators adds complexity to reconstructing the evolutionary origin of animals, Curr. Biol., 2020a, vol. 30, no. 22, pp. 4500–4509.
Tikhonenkov, D.V., Strassert, J.F.H., Janouškovec, J., Mylnikov, A.P., Aleoshin, V.V., et al., Predatory colponemids are the sister group to all other alveolates, Mol. Phylogenet. Evol., 2020b, vol. 149, art. ID 106839.
Timling, I., Walker, D.A., Nusbaum, C., Lennon, N.J., and Taylor, D.L., Rich and cold: diversity, distribution and drivers of fungal communities in patterned-ground ecosystems of the North American Arctic, Mol. Ecol., 2014, vol. 23, no. 13, pp. 3258–3272.
Tokarev, Y.S., Huang, W.F., Solter, L.F., Malysh, J.M., Becnel, J.J., and Vossbrinck, C.R., A formal redefinition of the genera Nosema and Vairimorpha (Microsporidia: Nosematidae) and reassignment of species based on molecular phylogenetics, J. Invertebr. Pathol., 2020, vol. 169, art. ID 107279.
Torreira, E., Louro, J.A., Pazos, I., González-Polo, N., Gil-Carton, D., et al., The dynamic assembly of distinct RNA polymerase I complexes modulates rDNA transcription, eLife, 2017, vol. 6, art. ID e20832.
Transcription of Ribosomal RNA Genes by Eukaryotic RNA Polymerase I, Paule, M.R., Ed., Berlin: Springer-Verlag, 1998.
Trzebny, A., Slodkowicz-Kowalska, A., Becnel, J.J., Sanscrainte, N., and Dabert, M., A new method of metabarcoding Microsporidia and their hosts reveals high levels of microsporidian infections in mosquitoes (Culicidae), Mol. Ecol. Resour., 2020, vol. 20, pp. 1486–1504.
Usachev, K.S., Yusupov, M.M., and Validov, S.Z., Hibernation as a stage of ribosome functioning, Biochemistry (Moscow), 2020, vol. 85, no. 11, pp. 1434–1442.
Vivier, E., Étude, au microscope électronique, de la spore de Metchnikovella hovassei n. sp.: appartenance des Metchnikovellidae aux Microsporidies, C. R. Hebd. Séan. Soc. Biol., 1965, vol. 260, no. 26, pp. 6982–6984.
Vossbrinck, C.R. and Woese, C.R., Eukaryotic ribosomes that lack a 5.8S RNA, Nature, 1986, vol. 320, no. 6059, pp. 287–288.
Vossbrinck, C.R., Maddox, J.V., Friedman, S., Debrunner-Vossbrinck, B.A., and Woese, C.R., Ribosomal RNA sequence suggests Microsporidia are extremely ancient eukaryotes, Nature, 1987, vol. 326, no. 6111, pp. 411–414.
Wadi, L. and Reinke, A.W., Evolution of microsporidia: an extremely successful group of eukaryotic intracellular parasites, PLoS Pathog., 2020, vol. 16, no. 2, art. ID e1008276.
Wagner, A., Energy constraints on the evolution of gene expression, Mol. Biol. Evol., 2005, vol. 22, no. 6, pp. 1365–1374.
Wakeman, K.C., Molecular phylogeny of marine gregarines (Apicomplexa) from the Sea of Japan and the Northwest Pacific including the description of three novel species of Selenidium and Trollidium akkeshiense n. gen. n. sp., Protist, 2020, vol. 171, no. 1, art. ID 125710.
Warner, J.R., The economics of ribosome biosynthesis in yeast, Trends Biochem. Sci., 1999, vol. 24, no. 11, pp. 437–440.
Weber, S.D., Hofmann, A., Wanner, G., Pilhofer, M., Agerer, R., et al., The diversity of fungi in aerobic sewage granules assessed by 18S rRNA gene and ITS sequence analyses, FEMS Microbiol. Ecol., 2009, vol. 68, no. 2, pp. 246–254.
Weiser, J., Contribution to the classification of microsporidia, Vestn. Cesk. Spol Zool., 1977, vol. 41, no. 4, pp. 308–321.
White, M.M., James, T.Y., O’Donnell, K., Cafaro, M.J., Tanabe, Y., and Sugiyama, J., Phylogeny of the Zygomycota based on nuclear ribosomal sequence data, Mycologia, 2006, vol. 98, no. 6, pp. 872–884.
Whiting, M.F., Carpenter, J.C., Wheeler, Q.D., and Wheeler, W.C., The Strepsiptera problem: phylogeny of the holometabolous insect orders inferred from 18S and 28S ribosomal DNA sequences and morphology, Syst. Biol., 1997, vol. 46, no. 1, pp. 1–68.
Wijayawardene, N.N., Hyde, K.D., Al-Ani, L.K.T., Tedersoo, L., Haelewaters, D., et al., Outline of Fungi and fungus-like taxa, Mycosphere, 2020, vol. 11, no. 1, pp. 1060–1456.
Wilms, R., Sass, H., Köpke, B., Köster, J., Cypionka, H., and Engelen, B., Specific bacterial, archaeal, and eukaryotic communities in tidal-flat sediments along a vertical profile of several meters, Appl. Environ. Microbiol., 2006, vol. 72, no. 4, pp. 2756–2764.
Wurzbacher, C., Rösel, S., Rychła, A., and Grossart, H.P., Importance of saprotrophic freshwater fungi for pollen degradation, PLoS One, 2014, vol. 9, no. 4, art. ID e94643.
Wuyts, J., De Rijk, P., van de Peer, Y., Pison, G., Rousseeuw, P., and De Wachter, R., Comparative analysis of more than 3000 sequences reveals the existence of two pseudoknots in area V4 of eukaryotic small subunit ribosomal RNA, Nucleic Acids Res., 2000, vol. 28, no. 23, pp. 4698–4708.
Wuyts, J., van de Peer, Y., and De Wachter, R., Distribution of substitution rates and location of insertion sites in the tertiary structure of ribosomal RNA, Nucleic Acids Res., 2001, vol. 29, no. 24, pp. 5017–5028.
Zhou, X., Montalva, C., and Arismendi, N., Neozygites linanensis sp. nov., a fungal pathogen infecting bamboo aphids in southeast China, Mycotaxon, 2017, vol. 132, no. 2, pp. 305–315.
Zuker, M., Mfold web server for nucleic acid folding and hybridization prediction, Nucleic Acids Res., 2003, vol. 31, no. 13, pp. 3406–3415.
ACKNOWLEDGMENTS
The authors are grateful to Prof. F.M. Freimozer for the kind advice.
Funding
The authors are grateful to the Russian Science Foundation for the support of the study of cDNA libraries of eugregarines, archigregarines, and blastogregarines (project no. 18-14-00123) and to the Russian Foundation for Basic Research for support of the work on determination of the diversity and phylogeny of unicellular consumers of unicellular organisms (project no. 18-04-01210).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests. The authors declare that they have no conflicts of interest.
Statement on animal welfare. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Additional information
Translated by A. Barkhash
Rights and permissions
About this article
Cite this article
Mikhailov, K.V., Nassonova, E.S., Shɨshkin, Y.A. et al. Ribosomal RNA of Metchnikovellids in Gregarine Transcriptomes and rDNA of Microsporidia Sensu Lato in Metagenomes. Biol Bull Rev 12, 213–239 (2022). https://doi.org/10.1134/S2079086422030069
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S2079086422030069