Both high and low dispersal? Apparently contradictory genetic patterns in the Antarctic littorinid gastropod Laevilacunaria antarctica
- 1Instituto de Ciencias Marinas y Limnológicas (ICML), Facultad de Ciencias, Universidad Austral de Chile, Valdivia, Chile
- 2Instituto Milenio Biodiversidad de Ecosistemas Antárticos y Subantárticos (Mi-BASE), Las Palmeras, Ñuñoa, Santiago, Chile
- 3Centro Fondap de Investigación en Dinámica de Ecosistemas Marinos de Altas Latitudes (IDEAL), Universidad Austral de Chile, Valdivia, Chile
- 4Departamento de Biología Marina, Facultad de Ciencias del Mar, Universidad Católica del Norte, Coquimbo, Chile
- 5Instituto Milenio en Socio-ecología Costera (SECOS), Coquimbo, Chile
- 6Cape Horn International Center (CHIC), Puerto Williams, Chile
- 7Centro de Investigación Gaia-Antártica, Universidad de Magallanes, Punta Arenas, Chile
- 8Centro i-mar, Universidad de los Lagos, Puerto Montt, Chile
- 9Laboratorio de Ecosistemas Antárticos y Subantárticos (LEMAS), Universidad de Magallanes, Punta Arenas, Chile
- 10Department of Zoology, University of Otago, Dunedin, New Zealand
- 11Laboratorio de Ecología Molecular (LEM), Departamento de Ciencias Ecológicas, Universidad de Chile, Las Palmeras, Ñuñoa, Santiago, Chile
How the near-shore marine benthic communities survived Quaternary glaciations in Antarctica is a major question for Southern Ocean biogeographers. Several hypotheses that consider life-history traits, such as bathymetric ranges and developmental modes, have been proposed. Near-shore species with high dispersive potential are expected to show star-like genealogies, with broadly distributed haplotypes surrounded by closely related variants at low frequencies, a consequence of rapid population post-glacial expansion mediated by larvae. By contrast, shallow-water taxa with low dispersal potential are expected to evidence marked genetic structure or even consist of separate cryptic species, due to their survival in small refugial populations subject to repeated bottlenecks. In this study we performed phylogeographic analyses on the high-latitude littorinid Laevilacunaria antarctica across its distribution from maritime Antarctica to South Georgia. This species is a highly abundant near-shore gastropod, commonly found on macroalgae, with low autonomous vagility and exhibits benthic protected development with no free-living dispersive stages. Such characteristics make this species a suitable model to test the above hypotheses. Contrary to low dispersal potential predictions, L. antarctica comprises a single unstructured unit across its distribution in maritime Antarctica, exhibiting a typical high-dispersal star-like, short-branched genealogy centered on a dominant broadly distributed haplotype. The South Georgia population, however, consists of a separate genetic unit, strongly differentiated from those of the maritime Antarctic. We estimate that these two genetic groups separated ~1.2 My, long before the Last Glacial Maximum and evolved independently in these areas. Our results thus provide partial support for the expected pattern for a near-shore marine benthic species with low innate dispersive potential. On a wider geographic scale (maritime Antarctica vs South Georgia), our results in L. antarctica provide support for the expected pattern for a near-shore marine benthic species with low innate dispersive potential. However, on a narrower scale (maritime Antarctica) life-history characters including the close association of the species with macroalgae, would allow dispersal and a species with low dispersal potential exhibits paradoxically an absence of genetic structure across hundreds of kilometers probably mediated by rafting.
Introduction
How the shallow benthic marine biota endured Quaternary glaciations, particularly the Last Glacial Maximum (LGM), is a major question in Antarctic biogeography (Barnes et al., 2006; Thatje et al., 2008; Allcock and Strugnell, 2012; Fraser et al., 2014; Riesgo et al., 2015; Chenuil et al., 2018; Crame, 2018; Lau et al., 2020; Baird et al., 2021). Ice advances across the Antarctic continental platform generated major landscape shifts and a drastic reduction of available habitats (Zachos et al., 2001; Pollard and DeConto, 2009; Fraser et al., 2012; González-Wevar et al., 2012; González-Wevar et al., 2013; McCay et al., 2016; Halanych and Mahon, 2018);. Accordingly, near-shore marine benthic communities would have been especially vulnerable as continental ice sheets extended over most of the narrow Antarctic shelf to about 500 m in depth (Ingólfson, 2004; Thatje et al., 2005; Barker et al., 2007; Dambach et al., 2012).
Quaternary glaciations severely impacted the abundance, demographic dynamics, structure, and spatial distribution of Antarctic (Convey et al., 2009; McGaughran et al., 2011; Fraser et al., 2012; González-Wevar et al., 2013; Riesgo et al., 2015; Chenuil et al., 2018; Guillemin et al., 2018; Halanych and Mahon, 2018; Maturana et al., 2020; Cakil et al., 2021; Levicoy et al., 2021) and sub-Antarctic (Waters, 2008; Fraser et al., 2009; Nikula et al., 2010; González-Wevar et al., 2011; González-Wevar et al., 2012; Cumming et al., 2014; Moon et al., 2017; González-Wevar et al., 2018; González-Wevar et al., 2021) near-shore marine organisms. Moreover, ice advances during glacial maxima would have enhanced speciation, particularly in species with nonpelagic developmental modes (direct developers), as populations became fragmented into small and isolated refugia across the Antarctic continental shelf (Brey et al., 1996; Pearse et al., 2009; Chenuil et al., 2018). Following this, interglacial periods generated the expansion of newly available habitats where populations and species would have expanded their distributions (Provan and Bennett, 2008; Marko et al., 2010; González-Wevar et al., 2013). Under a basic expansion-contraction model of Pleistocene biogeography (Provan and Bennett, 2008), refugial populations are expected to harbor higher levels of intraspecific genetic diversity compared to those that were heavily impacted by ice and/or recently recolonized ones (Marko, 2004; Maggs et al., 2008; Marko et al., 2010). Moreover, in some cases glaciations have been associated with the radiation and speciation of different marine near-shore invertebrates (Thornhill et al., 2008; Wilson et al., 2009; Allcock et al., 2011; Baird et al., 2011; Baird et al., 2021), a process also known as ‘the Antarctic diversity pump’ (Clarke and Crame, 1989; Clarke and Crame, 1992; Chenuil et al., 2018).
In a review of genetic signatures induced by Quaternary events in Antarctic taxa, Allcock and Strugnell (2012) proposed that two life-history traits, bathymetric ranges and developmental modes, played a key role. Shallow species with dispersive potential would exhibit star-like haplotype networks, with broadly distributed common haplotypes and closely related variants occurring at low frequencies. Such a pattern is associated with rapid population expansion, probably mediated by larvae dispersion, and has been recorded in the sea urchin Sterechinus neumayeri (Díaz et al., 2011), the nemertean Parborlasia corrugatus (Thornhill et al., 2008), the Antarctic limpet (González-Wevar et al., 2013) and the shrimp Chorismus antarcticus (Raupach et al., 2010). By contrast, the genetic signature predicted for shallow species with low dispersal potential includes disjunct haplotypes networks comprising multiple smaller genetic units, indicative of small populations isolated in glacial refugia that underwent bottlenecks. Due to the absence of free dispersive stages, recolonization is slow, allowing time for genetic drift and possible adaptive differentiation before secondary contact of isolated populations. Examples of taxa displaying this pattern include the amphipods Eusirus (Baird et al., 2011), the pycnogonid Colossendeis megalonyx (Krabbe et al., 2010), cephalopods (Allcock et al., 2011), and the nudibranch Doris kerguelensis (Wilson et al., 2009).
The high-latitude littorinid species Laevilacunaria antarctica (Martens van and Pfeffer, 1886) is one of the most abundant inter- and subtidal gastropods on macro-algae and rocky shores across ice-free areas of the Antarctic Peninsula and sub-Antarctic island of South Georgia (Cantera and Arnaud, 1984; Iken, 1999; Amsler et al., 2015; Martin et al., 2016; Amsler et al., 2019; Amsler et al., 2022; Schmider-Martínez et al., 2023). As with other Laevilitorininae genera such as Laevilitorina (Simpson and Harrington, 1985), L. antarctica exhibits low autonomous vagility, benthic protected development with the absence of free-living dispersive stages, a mode or reproduction where crawling juveniles emerge directly from egg masses. Generally, high-latitude littorinids are considered to be poor dispersers, with most species having narrow distributions restricted to particular areas of the Southern Ocean (González-Wevar et al., 2022; Rosenfeld et al., 2023). Based on its broad distribution and abundance across the maritime Antarctica, its narrow bathymetric range, and its reproductive mode, L. antarctica represents a suitable model to test the hypotheses proposed for Antarctic Quaternary biogeography of shallow marine benthic organisms.
In this study, we performed population-based molecular analyses of Laevilacunaria antarctica across its known distribution. We included more than 320 specimens collected from across the Antarctic Peninsula (AP) and the nearby South Shetland Islands (SSI), as well as from sub-Antarctic South Georgia (SG). Comparative mitochondrial (COI) and nuclear (28S rRNA) genetic analyses allowed us to unravel the legacy of Quaternary glaciations in the patterns of genetic diversity and structure of this species. Considering the lack of larval dispersive stages in the species and its narrow bathymetric range, we predicted that L. antarctica would display several disjunct haplotype networks separated into smaller genetic units, as recorded in other Antarctic species with similar life history traits. Through this research we aimed to understand how key elements of the near-shore Antarctic marine benthic communities endured Quaternary climate shifts associated with glacial and interglacial periods.
Materials and methods
Sample collection, DNA extraction and amplifications
Specimens of L. antarctica were collected by hand and by scuba divers from intertidal and subtidal rocky-shore ecosystems between 2015 and 2021 during the Chilean Scientific Antarctic Expeditions (ECAs) across the species distribution in the maritime Antarctica (AP and SSI) and SG (Figure 1; Table 1). The identification of specimens was done following the original descriptions and revisions (Martens van and Pfeffer, 1886; Smith, 1879; Preston, 1916; Powell, 1951) as well as recent literature (Arnaud and Bandel, 1979; Cantera and Arnaud, 1984; Engl, 2012). Individuals were fixed in situ using 95% ethanol and transported for further molecular analyses. All the specimens were photographed and measured for future comparative morphological studies. Preparation of DNA from the whole animal used the standard salting-out methodology (Aljanabi and Martinez, 1997) and the QIAGEN DNEasy Blood & Tissue kit (QIAGEN Inc.). We amplified a partial fragment of the mitochondrial cytochrome c subunit I gene (COI) using universal primers LCO1490 and HCO2198 (Folmer et al., 1994). Similarly, we used the universal primers 900F and 1600R (Littlewood et al., 2000) to amplify a fragment of the nuclear 28S rRNA gene. PCR amplicons were purified and sequenced in both directions at Macrogen Inc (Seoul, South Korea). Forward and reverse sequences were assembled and edited for each marker independently using GENEIOUS (http://www.geneious.com). Alignments and base composition of nucleotide sequences analyses were done for each marker in MUSCLE (Edgar, 2004) in MEGA 11 (Tamura et al., 2021), respectively. Wright’s mitochondrial codon usage was computed using the Effective Number of Codons (ENC) in DnaSP v5 (Librado and Rozas, 2009). The data presented in the study are deposited in the GenBank repository, accession number COI (ORT701885 - ORT701931) and 28S rRNA (ORT711532 - ORT711533).
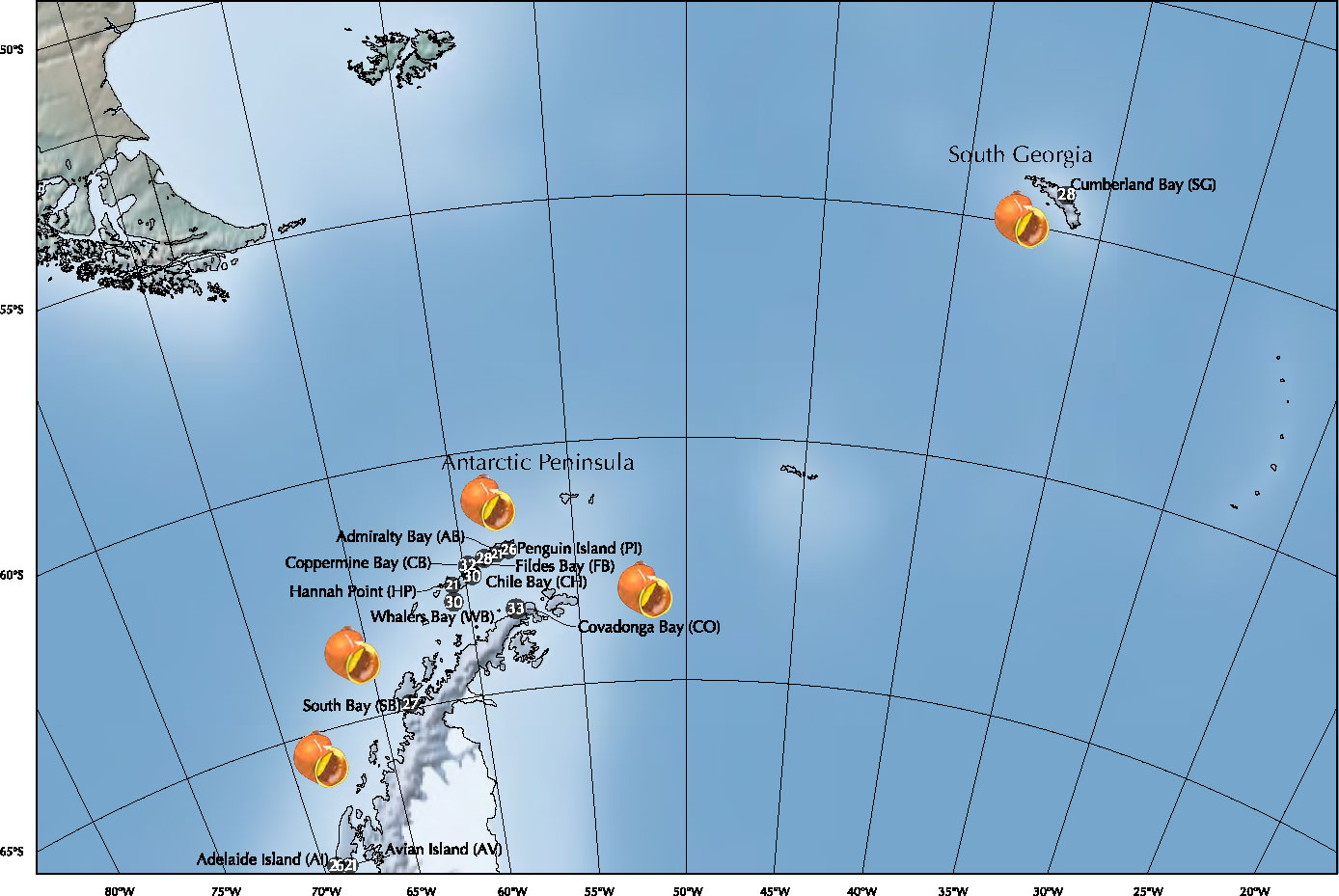
Figure 1 Sampling localities of Laevilacunaria antarctica populations across its distribution in maritime Antarctica (SSI/AP) and South Georgia (SG).
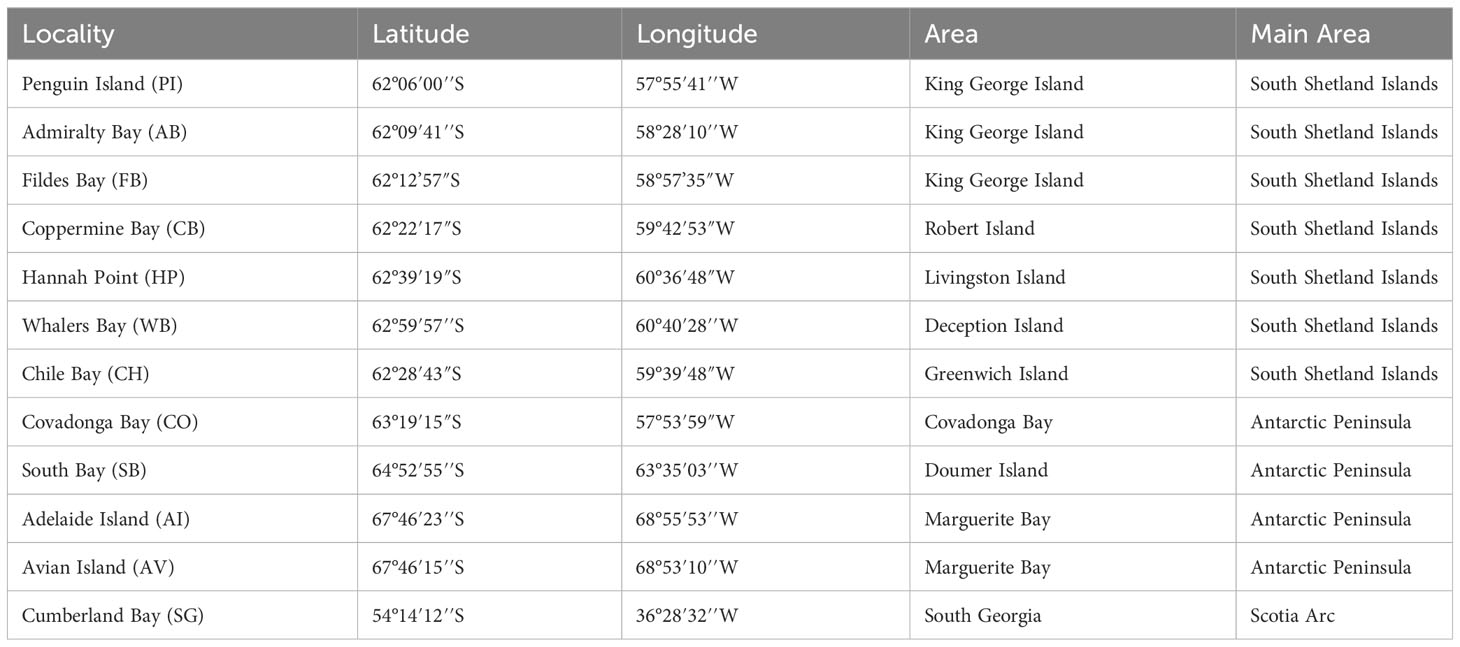
Table 1 Sampled localities of Laevilacunaria antarctica across its distribution in maritime Antarctica (MP) and South Georgia.
Genetic diversity and population structure in L. antarctica
We performed a DNA saturation analysis following Xia and Xie (2001) to evaluate how transitions accumulate in relation to nucleotide divergence in the complete L. antarctica COI data set. We then estimated levels of population mtDNA polymorphism through standard diversity indices including number of haplotypes (k), haplotype diversity (H), number of segregating sites (S) and the number of private haplotypes (p. hap) per locality. Moreover, we also determined the average number of pairwise differences (П) and the nucleotide diversity (π) across the species distribution using DnaSP.
Patterns of population differentiation were estimated following Pons and Petit (1996), using haplotype frequencies (GST) and mean pairwise differences (NST) in Arlequin v.3 (Excoffier et al., 2005). The statistical significance of genetic pairwise differences was calculated using permutation tests (20,000 iterations) and the adjustment for multiple testing was done through False Discovery Rate (FDR) correction (Narum, 2006). Moreover, we estimated levels of genetic differentiation using the nearest-neighbor statistic (Snn), which measures how often nearest-neighbor (in sequence space) sequences are from the same locality in geographic space (Hudson, 2000). The statistical significance of Snn was determined using a permutation test (20,000 iterations).
We used two different clustering methods to determine the spatial genetic structure of L. antarctica. First, we determined the number and the composition of panmictic groups and the spatial boundaries using a Bayesian model computed in GENELAND v.2.0.0 (Guillot et al., 2005) in the R environment (Ihaka and Gentleman, 1996). This analysis implements a Markov Chain Monte Carlo (MCMC) procedure to estimate the best clustering of samples considering genetic and geographic information. Analyses were run using 50 x 106 MCMC iterations sampled every 1,000 steps. Assembled scores were graphed against generations in Tracer v.1.5. (Rambaut et al., 2018) to identify stationarity and the number of generations to be discarded as burn-in. A maximum number of clusters (K = 13) were run to estimate the model parameters and posterior probabilities of group membership. Second, we determined the spatial genetic structure in L. antarctica by estimating the number and composition of groups that were most differentiated based on sequence data set using Spatial Analysis of Molecular Variance (SAMOVA) (Dupanloup et al., 2002). This analysis partitions the genetic variance into i) within populations, ii) among populations within groups and iii) among groups.
Demographic analyses in L. antarctica
Haplotype genealogical relationships in L. antarctica populations were reconstructed using median-joining and maximum-parsimony networks in Network 10 (Forster et al., 2001) and Hapview (Salzburger et al., 2011), respectively. Moreover, we performed neutrality statistical tests (Tajima’s D and Fu’s FS) using DnaSP for the whole COI data set, for each recognized group, and for each locality to estimate whether sequences deviate from mutation-drift equilibrium. Population demographic histories were estimated comparing the distribution of pairwise differences between haplotypes (mismatch distribution) for each recognized group to the expected distribution under the sudden expansion growth model of Rogers and Harpending (1992). Finally, for comparative purposes, we also constructed genealogical relationships in L. antarctica using sequences of the nuclear marker 28S rRNA.
We reconstructed past population dynamics through time in the recognized L. antarctica’s genetic clusters using a Bayesian skyline plot method implemented in BEAST v.1.10.4 (Drummond and Rambaut, 2007; Drummond et al., 2012). Three evolutionary models (strict clock, uncorrelated lognormal and uncorrelated relaxed clock) were tested and compared statistically using a Bayes factor test (Suchard et al., 2001) in Tracer and the strict clock model was the best fit for each of the COI data-set clusters. Following this, we performed three independent Bayesian MCMC runs for 250 x 106 generations (sampled every 1000 iterations), using the GTR + I + Γ model, previously estimated in MrModeltest v.2.3 (Nylander, 2004). Molecular evolutionary studies have yielded a wide range of rate estimates for different genes and taxa. Several studies based on population-level and intraspecific data have generated remarkably high estimates of mutation rate, which contrast with substitution rates inferred in phylogenetic studies. Consequently, BSP analyses were done using a tenfold evolutionary rate estimated for littorinids (Reid et al., 2012). Such a rate correction was assumed considering the time-dependence of molecular rate proposed by Ho et al (2005; 2007; 2011). The first 10% of the parameter values were discarded as a burn-in and the convergence of runs was confirmed with Tracer, ensuring a minimum of 1000 effective sampling for each statistic (ESSs > 1000).
Results
Genetic diversity in L. antarctica
The whole COI data set in L. antarctica included 323 specimens (SG = 28; SSI = 188; AP = 107) and comprised 687 nucleotide positions coding for 229 amino acids. No insertion/deletions or stop codons were detected. Moreover, mitochondrial (mtDNA) and nuclear (nucDNA) sequences were not saturated and no evidence of mtDNA codon bias was found (ENC = 46.75). Four amino-acid substitutions (positions 11, 44, 144, and 171) were recorded in L. antarctica using the invertebrate mitochondrial table. Low levels of genetic diversity characterized populations of L. antarctica, with 48 polymorphic characters (6.9%) and 29 of them (60.4%) were parsimony informative. Sequences were A – T rich (67.2%). The haplotype diversity (H) varied between 0.123 (Robert Island, SSI) and 0.675 (Cumberland Bay, SG) (Table 2). The number of polymorphic sites (S) varied between 1 (Admiralty Bay, SSI) and 10 (Cumberland Bay, SG). Similarly, the number of haplotypes/private haplotypes varied among the analyzed localities between 2/0 (Admiralty Bay, SSI) and 11/11 (Cumberland Bay, SG) (Table 2). Finally, the average number of nucleotide differences (П) and the mean nucleotide diversity (π) were very low in most localities across the maritime Antarctica (SSI and AP), whereas the diversity of these indices was higher in SG (Table 2). The mean level of mtDNA genetic divergence between SSI/AP and SG populations of L. antarctica was 1,74%. Levels of nucDNA diversity were much lower in the species, SSI and AP specimens (n = 123) shared the same allele which was differentiated by two mutation steps from the sequences recorded at South Georgia.
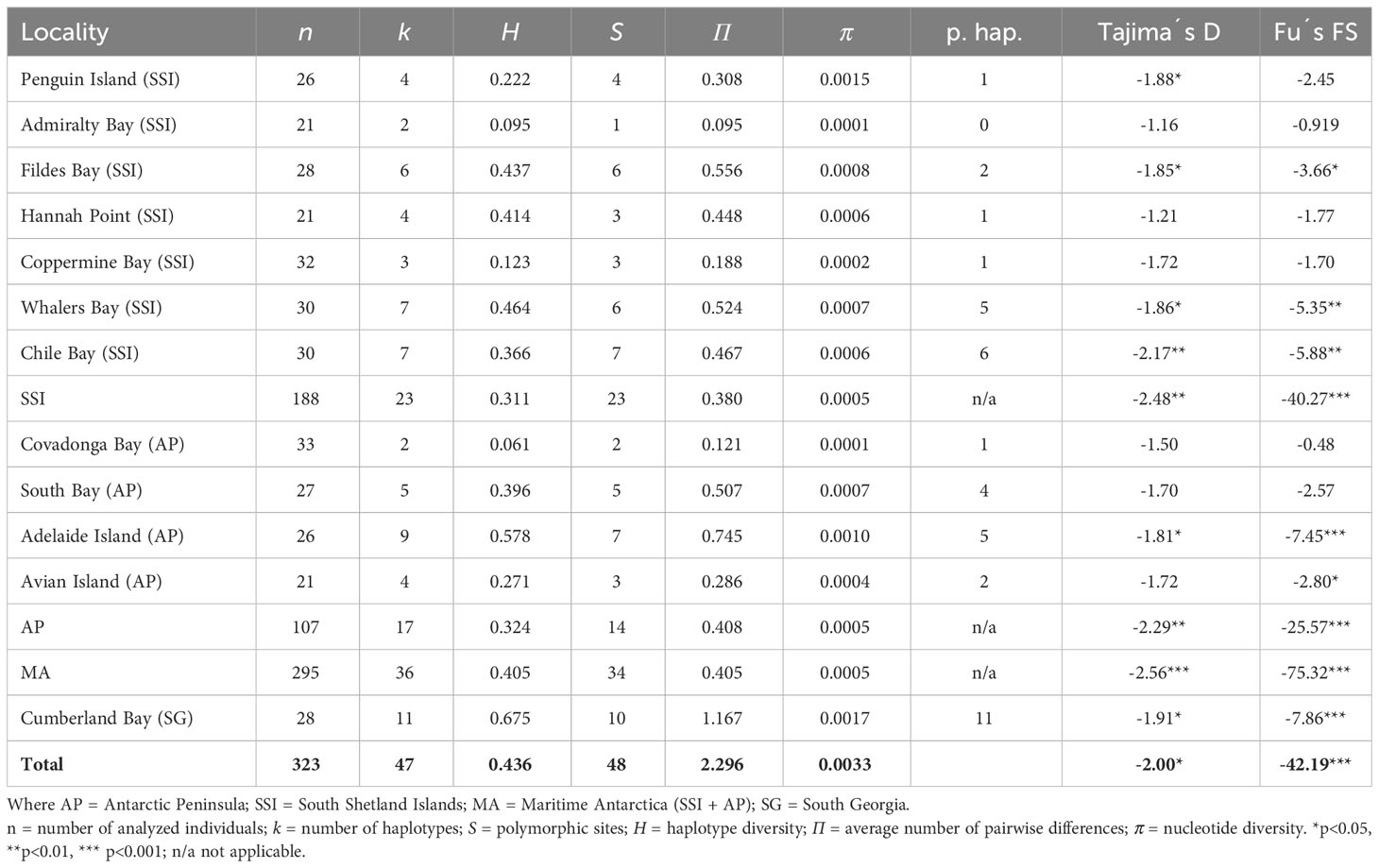
Table 2 Diversity indices, private alleles, and neutrality tests in Laevilacunaria antarctica populations across its distribution in maritime Antarctica and South Georgia.
Genetic structure in L. antarctica
Mean general values of differentiation estimated were low, especially considering average GST (0.112) and NST (0.170). In fact, general levels of differentiation among maritime Antarctic populations were extremely low, considering average GST (0.043) and NST (0.034). Pairwise population comparisons did not recognize significant structure among maritime Antarctica populations (Table 3). Nevertheless, some populations showed significant differences after FDR corrections. By contrast, GST and NST comparisons recorded significant differences between SG and the rest of the analyzed populations from the maritime Antarctica (Table 3).
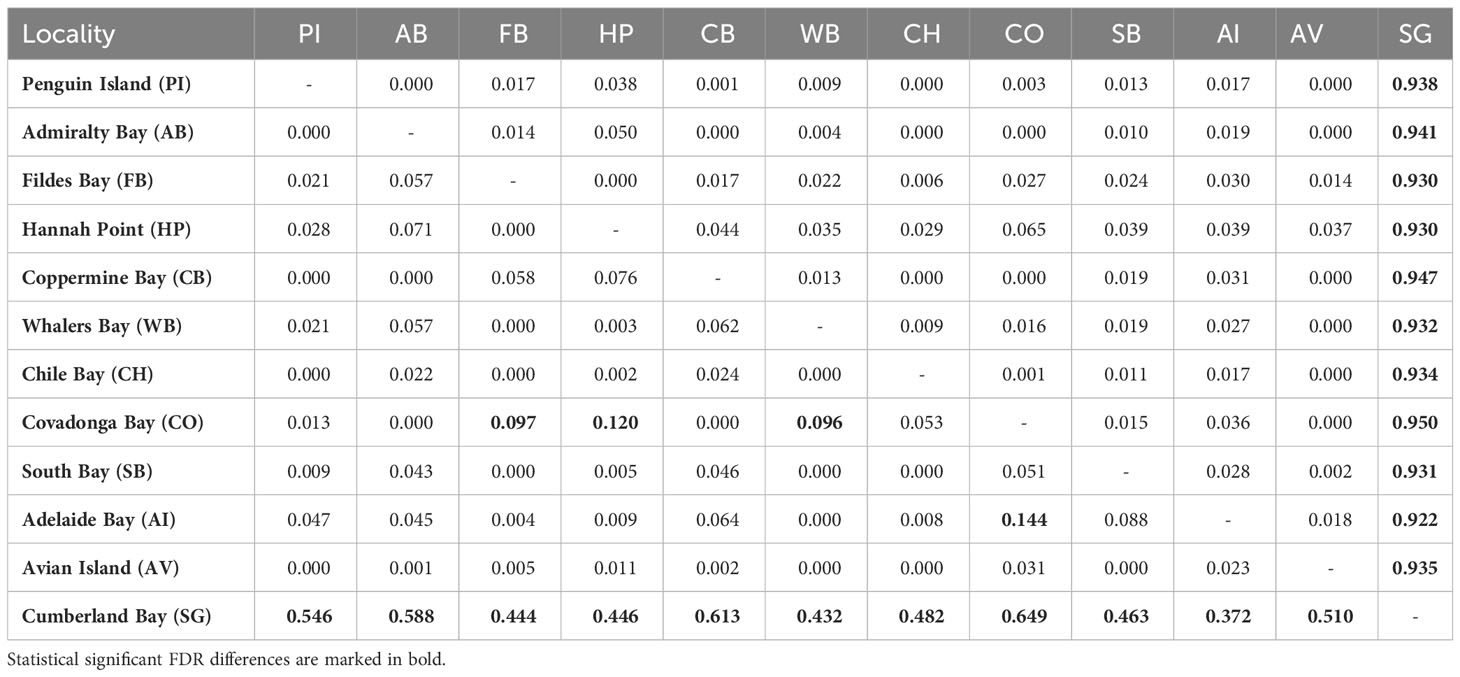
Table 3 Pairwise GST (below the diagonal) and NST (above the diagonal) values calculated among the analyzed populations of Laevilacunaria antarctica. 20,000 iterations.
The nearest neighbor statistic in L. antarctica (Snn = 0.199) showed low but significant levels of phylogeographic signal (p < 0.0001). Nevertheless, when this analysis was carried out considering the main pattern of genetic differentiation recorded in the species (maritime Antarctic vs South Georgia), Snn became extremely high (Snn = 1.00) and significant, showing the high degree of phylogeographic signal found between maritime Antarctica and SG. The pattern of genetic structure was supported by the model based on the Bayesian clustering algorithm, which detected two main clusters (K = 2). Again, the first cluster included all localities from the maritime Antarctica (Figure 2A), while the second one comprised only South Georgia (Figure 2B). Values of cluster membership were very high (P = 1.000) for all the individuals and the mean probability value (P = 0.5) corresponds to the boundary between these clusters and runs across the Scotia Ridge located between South Georgia and the Antarctic Peninsula. Similarly, SAMOVA analyses detected two maximally differentiated genetic groups – maritime Antarctica and South Georgia – accounting for 59.35% of the total variance, in comparison with only 0.83% due to within-group variation among localities (Table 4). Considering the level of mtDNA genetic divergence (1.74%) and the strong phylogeographic signal recorded between SSI/AP and SG we performed divergence time estimations analysis under a phylogenetic framework. For this a relaxed molecular-clock using an uncorrelated-lognormal (ucld) model of molecular evolutionary rate heterogeneity and the GTR + I + G model of substitution was implemented in BEAST v.1.7.5 (Drummond & Rambaut, 2007; Drummond et al., 2012). Four chains were run for 200 x 106 generations and trees were sampled every 10,000 generations. For divergence time estimations we include in the analyses several calibration points within the evolution of Littorinidae estimated for the most common ancestors (TRMCA) of the genera Echinolittorina, Littorina, Littoraria, Austrolittorina, Tectarius and Afrolittorina) following Reid et al. (2012), and with special emphasis on higher latitude groups (González-Wevar et al., 2022). According to our divergence-time analyses the separation between SSI/AP and SG genetic clusters occurred around 1.1 Ma (2.0 – 0.55 Ma).
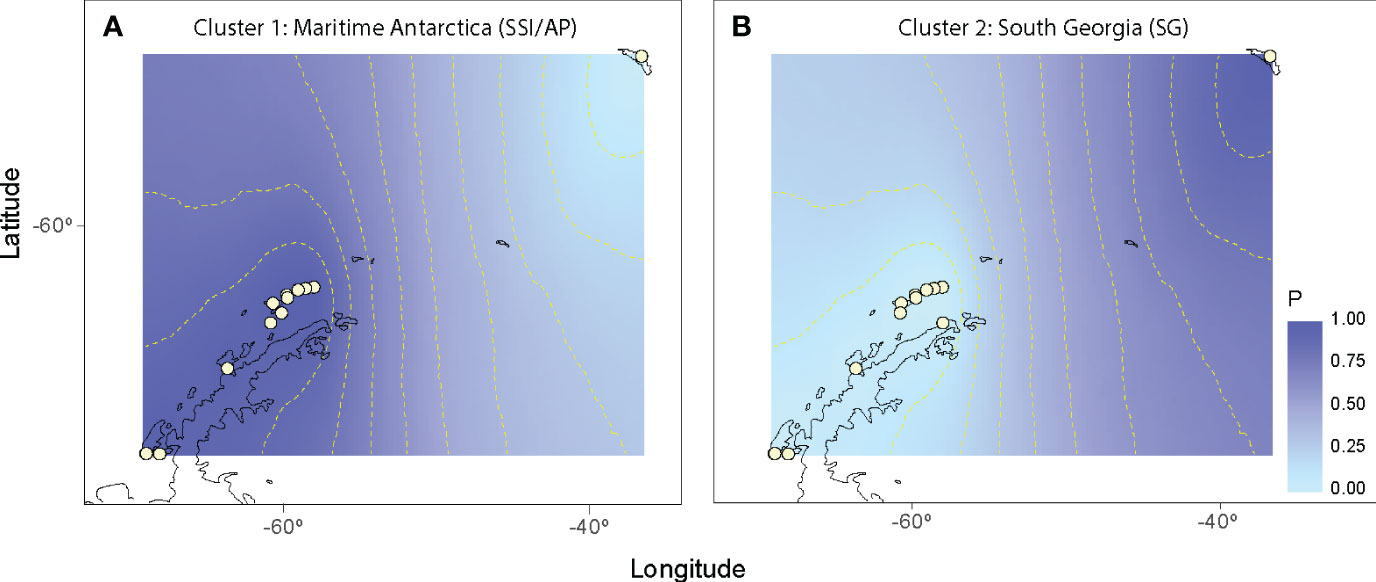
Figure 2 Spatial output from Geneland using Laevilacunaria antarctica populations. White circles indicate the relative positions of the sampling localities across maritime Antarctica and South Georgia. Darker and lighter shadings are proportional to posterior probabilities of membership to particular genetic cluster (A = maritime Antarctica and B = South Georgia). Posterior probabilities were plotted against the shapefile of the Antarctic coastline available in GEOdas (NOAA) and filtered using GEOdas Coastline Extractor v.1.1.3.1 (https://www.ngdc.noaa.gov/mgg/geodas/geodas.html).
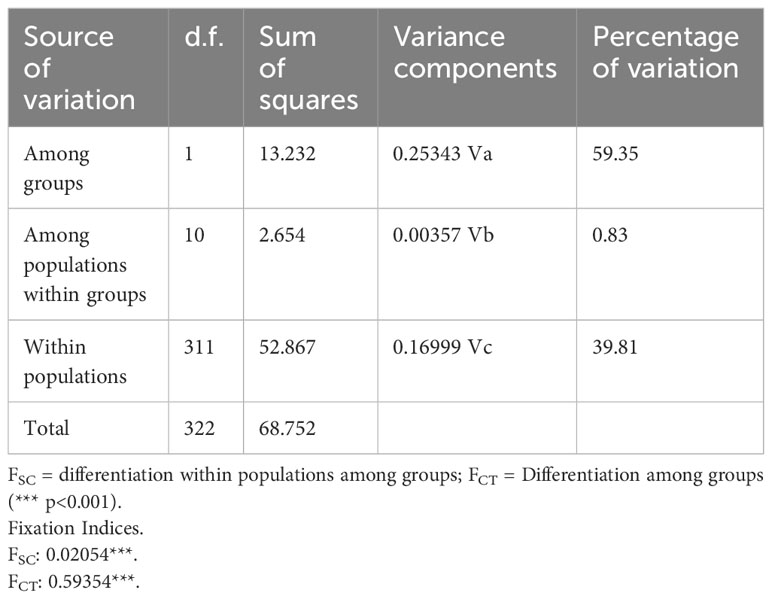
Table 4 Spatial Analysis of Molecular Variance (SAMOVA) depicting the percentage of variation explained among groups (maritime Antarctica and South Georgia), among populations within groups and within populations.
Demographic reconstructions
The parsimony mtDNA network of L. antarctica included 47 haplotypes and clearly discriminated two main groups: maritime Antarctica and South Georgia, separated by nine substitutional steps (Figure 3A). Both maritime Antarctica and South Georgia clusters exhibited typical starlike topologies and short genealogies. In maritime Antarctica, the central haplotype (H01) was the most frequent (82%), broadly distributed and surrounded by 35 haplotypes of low frequency (fewer than four individuals). A second dominant haplotype (H02) was found in 57% of the SG individuals and was surrounded by ten closely derived ones (Figure 3A). As expected, for star-like genealogies, global Tajima’s D and Fu’s neutrality tests were negative and significant for each recognized cluster and for the whole COI data set (Table 2). The distribution of pairwise differences depicted for the maritime Antarctica showed a typical L-shaped distribution. In contrast, the distribution of pairwise differences for South Georgia was bimodal. As stated above, nucDNA parsimony network showed that the maritime Antarctica populations specimens exhibited the same allele (HI) while SG specimens carried another one (HII) separated by two mutational steps (Figure 3B).
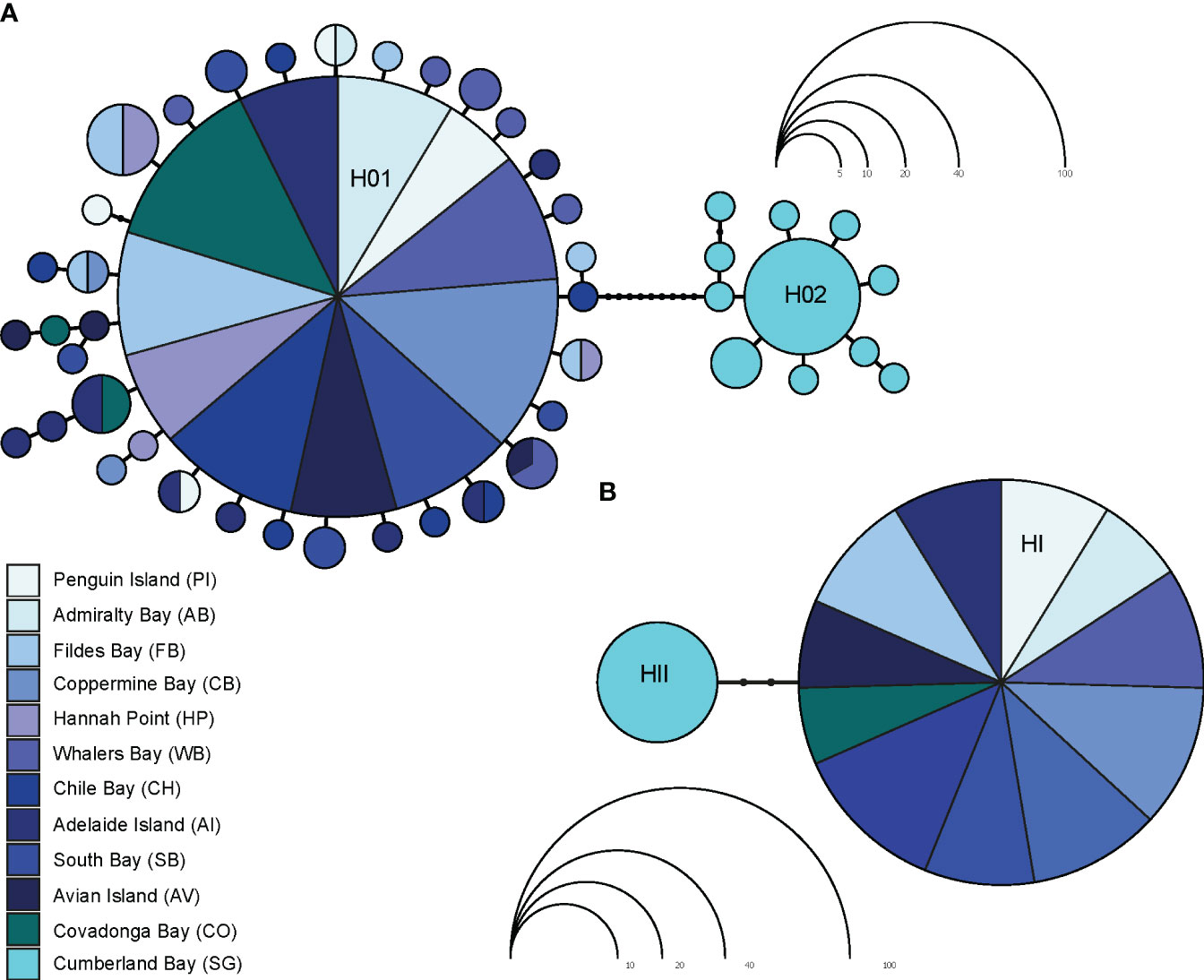
Figure 3 Parsimony mtDNA (A) and nucDNA (B) networks in Laevilacunaria antarctica across its distribution in maritime Antarctica and South Georgia. Each haplotype/allele is represented by a colored circle indicating the locality. The size the haplotypes/alleles is proportional to their frequencies.
Bayesian skyline-plot analyses identified similar trajectories in terms of the time since the most recent common ancestors (trmca) and populations expansions between maritime Antarctica and South Georgia (Figure 4). Population expansion of the maritime Antarctica populations occurred approximately 5,000 years ago while the expansion at South Georgia occurred around 9,000 years ago. The tmrca of SSI/AP occurred around 12,000 years ago while the tmrca for SG occurred 17,000 years ago (Figure 4).
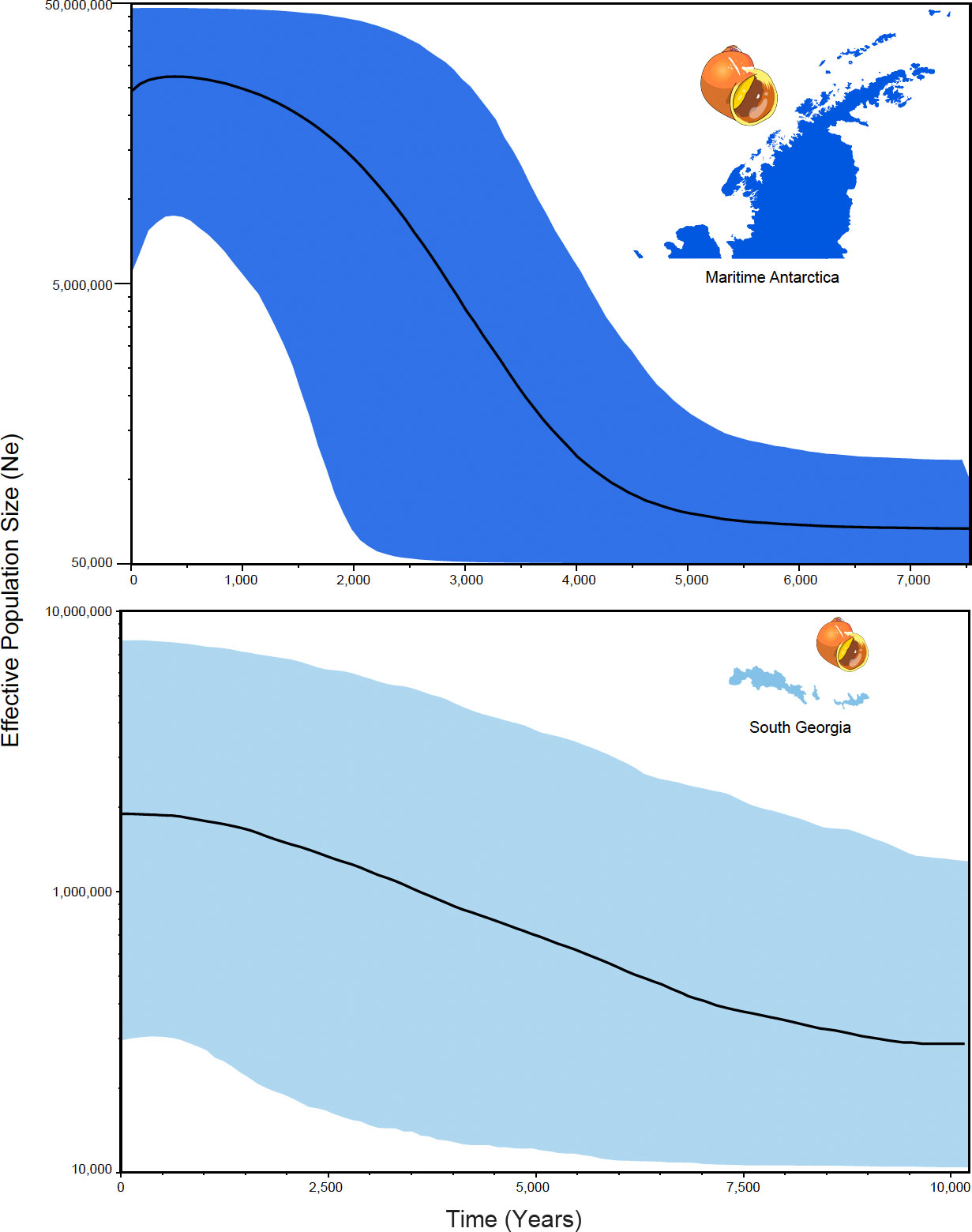
Figure 4 Historical demographic trajectories of the effective population sizes (Ne) estimated under a Bayesian skyline-plot approach based on COI sequences of L. antarctica. The y-axis represents the effective population size (Ne) while the x-axis shows the time since the present. The median estimates are shown for each genetic cluster recorded in the species (maritime Antarctica and South Georgia).
Discussion
Our results show that Laevilacunaria antarctica has a remarkably low level of genetic diversity across much of its known range, with the presence of a single dominant COI haplotype in our sampled populations from Penguin Island (62°06’), the farthest north locality in the South Shetland Islands, to the southernmost point in Avian Island (67°46’) in the Antarctic Peninsula. There is marginally more diversity further afield, in South Georgia, although again one haplotype, albeit a very different one, dominates. The only population structure is between South Georgia and the remaining populations, with no shared haplotypes and at least nine substitutional steps between haplotypes from these two parts of the species’ range. Effectively, the haplotype map is one of two star-like genealogies linked by a nine-step branch. Nevertheless, such results should be taken with caution, considering that we only included one locality from South Georgia. In this context, future samplings in South Georgia are required to increase our sampling effort across this island and corroborate the diversity pattern here recorded.
Our findings provide only limited support for the predictions of Allcock and Strugnell (2012), and indeed some evidence to the contrary. As Laevilacunaria lacks any pelagic dispersal stage, yet the low levels of diversity and the absence of genetic structure showed by star-like networks in the maritime Antarctica across hundreds of kilometers are more congruent with the pattern of genetic diversity these authors envisaged in highly dispersive species. Nevertheless, our analyses detected a marked phylogeographic signal between sub-Antarctic South Georgia and maritime Antarctic localities, fitting more closely with a poor dispersal capacity at large geographical scale. The absence of shared haplotypes, together with the number of fixed mutations (n = 9) between these areas are strong evidence of distinct genetic and evolutionary units in L. antarctica. Indeed, divergence-time estimations of these Significant Evolutionary Units (ESUs) suggest that they were separated ~ 1.1 million years ago (Ma), long before the LGM and, ever since, have evolved independently in these areas of the Southern Ocean.
How do we explain these apparently contradictory results? The absence of population structure in the maritime Antarctica matches previous molecular studies of near-shore marine benthic invertebrates such as the Antarctic limpet Nacella concinna (Hoffman et al., 2010; González-Wevar et al., 2013), the trochoid gastropod Margarella antarctica (González-Wevar et al., 2021), and the sea urchin Sterechinus neumayeri (Díaz et al., 2011; Díaz et al., 2018). Nacella and Sterechinus species possess a pelagic larval stage and such patterns are explained by their respective higher dispersal potentials. By contrast, Margarella and Laevilacunaria species exhibit benthic protected development, but they live closely associated with macroalgae on which they graze and reproduce. Accordingly, it is possible that Margarella and Laevilacunaria could maintain connectivity through rafting of adults and/or egg-masses attached to macroalgae. Alternatively, the low levels of genetic diversity and the absence of genetic structure in L. antarctica across the maritime Antarctica is probably associated to strong founder effects from bottlenecked refugial population. This same argument would apply to the South Georgian populations of L. antarctica. The lower frequency of buoyant macroalgae in the maritime Antarctic represented by Crystosphaera jacquinotti, however, presumably limits the extent of this rafting, precluding dispersal of L. antarctica (and, indeed, also occur in the trochoid M. antarctica) to the more distant areas such as South Georgia. Events of long-distance colonization by rafting have been documented among non-buoyant macroalgae (Fraser et al., 2013; McCay et al., 2016). Evidence of macroalgae fronds enclosed in drift ice have been reported at sea (Guillemin M-L pers. comm.) and these fronds have been recovered far away from the coast (Guillemin et al., 2018). In fact, across the same study are, several non-buoyant red algae also exhibit very low levels of genetic diversity and a complete absence of genetic structure (Guillemin et al., 2018). During Quaternary glaciations, the study area, SSI/AP and SG, have been located south of the Antarctic Polar Front (APF). Hence, the positition of the APF can be excluded as a potential barrier to gene flow between these areas. Drifter-based data have demonstrated that the Antarctic Circumpolar Current flows from the maritime Antarctica to South Georgia with a velocity that would allow to drift between these two areas around two months (Matschiner et al., 2009).
The lack of genetic variation in the maritime Antarctica, with one dominant haplotype in the centre of a star-like genealogy, fits well with the idea that L. antarctica survived a bottleneck induced by the LGM in one or (possibly) more shelf refugia. The slightly less star-like pattern, and the smoother historical demographic curve in South Georgia implies, perhaps, a less drastic reduction in population size and/or an earlier population expansion.
We note that the levels of nucleotide diversity recorded in L. antarctica are significantly lower than those found in temperate (Doellman et al., 2011; Silva et al., 2013; Sotelo et al., 2020; Blakeslee et al., 2021) and sub-Antarctic littorinids (González-Wevar et al., 2022). In fact, levels of genetic diversity in the species are much lower than those found in most other groups of Antarctic marine molluscs (Wilson et al., 2009; González-Wevar et al., 2013; Fassio et al., 2019; Levicoy et al., 2021) and fishes (Parker et al., 2002; Damerau et al., 2012; Damerau et al., 2014; Hüne et al., 2015). Nevertheless, several species of Antarctic near-shore marine invertebrates are characterized by low genetic diversity including the nemertean Parborlasia (Thornhill et al., 2008), the crustacean Chorismus antarcticus (Raupach et al., 2010), the echinoid Sterechinus neumayeri (Díaz et al., 2011) and the trochoid Margarella antarctica (González-Wevar et al., 2021). Drastic reduction of habitats has presumably affected populations of these Antarctic species and left characteristic signatures of drastic bottlenecks and/or founder effects (Aronson et al., 2007; Clarke and Crame, 2010; Allcock and Strugnell, 2012; González-Wevar et al., 2013; Riesgo et al., 2015; Guillemin et al., 2018).
Genetic structure
Our study adds to the diversity of biogeographic patterns seen in different taxa across maritime Antarctica and South Georgia in different taxa. This variation ranges from the absence of genetic differentiation, as in nototheniod fishes like Gobionotothen gibberifrons (Matschiner et al., 2009) and Notothenia rossi (Young et al., 2015), through marked phylogeographic structure, as in the limpet Nacella concinna (Hoffman et al., 2011; González-Wevar et al., 2013) and the notothenioid fish Champsocephalus gunnari (Young et al., 2015), to the presence of clearly divergent species-level clades, as in the gastropod Margarella (González-Wevar et al., 2021), the nudibranch Doris (Wilson et al., 2009), the crinoids of the genus Promachocrinus (Wilson et al., 2007), and the notothenioid genus Lepidonotothen (Dornburg et al., 2016).
The mtDNA (COI) levels of genetic divergence recorded between maritime Antarctic and South Georgian populations (1.74%) is lower than those recorded between clearly recognized littorinid species (Williams et al., 2003; Williams and Reid, 2004; Reid et al., 2012) and particularly in higher latitude groups (González-Wevar et al., 2022; Rosenfeld et al., 2023). Under this molecular-based criteria, we would not treat the SSI/AP and SG populations as different species. Appropriate names would be available, however: the type locality of L. antarctica is South Georgia, whereas Deception Island, one of the South Shetland Islands, is the type locality for L. bransfieldensis, considered a junior synonym by Simone (2018). Nevertheless, our results in Laevilacunaria could probably be interpreted as an incipient speciation process, as has been hypothesized for other groups of marine organisms including Nacella (González-Wevar et al., 2011; González-Wevar et al., 2017), octocorals (Dueñas et al., 2016), Doris (Wilson et al., 2009), Pareledone (Allcock et al., 2011), nothothenioid fishes (Near et al., 2012; Dornburg et al., 2016) and even penguins (Vianna et al., 2017; Frugone et al., 2019). To further understand such evolutionary and biogeographic hypotheses in Laevilacunaria, future studies through morphological (radular and geometric morphometrics) analyses are needed. Addressing this important question in Laevilacunaria will provide new insights concerning biogeographical and diversity patterns in this widespread maritime Antarctica gastropod group. Moreover, integrative systematic studies including phylogenetic, genomic and morphological analyses of the whole genus are also required to evaluate the potential role of vicariance and long-distance dispersal in the biogeography of Laevilacunaria. The inclusion of the poorly known Kerguelen species L. pumilio and the partially sympatric Antarctic species L. bennetti will allow us to determine the origin and diversification of Laevilacunaria in different provinces of the Southern Ocean. Finally, through geometric morphometric it will be possible to evaluate the relevance of cryptic speciation in the evolution of the group across the Southern Ocean, a region of the planet that has been relatively neglected in spite of its global relevance.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: GenBank under the following Accession Numbers: COI (ORT701885 - ORT701931) and 28S rRNA (ORT711532 - ORT711533).
Ethics statement
The animal study was approved by Comité de Ética, Facultad de Ciencias, Universidad Austral de Chile. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
CG: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. YP: Formal analysis, Investigation, Methodology, Software, Writing – original draft. NS: Formal analysis, Methodology, Software, Writing – review & editing. SR: Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. CM: Writing – original draft, Writing – review & editing. VJ: Formal analysis, Methodology, Writing – original draft. AS: Formal analysis, Methodology, Writing – original draft. KG: Writing – original draft. HS: Conceptualization, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing. EP: Conceptualization, Formal analysis, Methodology, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by Fondecyt Regular Project 1210787, ANID – Millenium Science Intitative program – ICN2021_002, INACH Project RG_18-17 and Program Fondap IDEAL (15150003) to CG-W. We also thank Fondecyt Postdoctoral Project 3210063 to CM and the program ANID/BASAL PR-06-CRN-18 (Universidad de Magallanes) to CM and SR.
Acknowledgments
Many thanks to Angie Díaz, Zambra López, Valentina Bernal, Jaime Ojeda, Cristian Ríos, Javier Naretto, Simon Morley and Paul Brickle for field assistance and samplings in maritime Antarctica and South Georgia. The authors thank the Chilean Antarctic Institute (INACH) and the Chilean Navy Aquiles and Oscar Viel Vessels for ECA53 – ECA55 and the crew of the Betanzos Ship during ECA58 and ECA59. We also would like to thank Commandant Charcot Cruise and the Scientist leader Geoffroy Dekersauson from Ponant Expeditions LEGCC120302023 and SEDNA/PONANT Expedition leg CC171222. We are grateful to the support of the South Atlantic Environmental Research Institute (SAERI) and the Shallow Marine Survey for collection of higher latitude littorinid specimens in South Georgia. At the same time, field access to South Georgia was supported by the British Antarctic Survey (BAS) and the collected specimens were authorized under the permit number 2017/058 to CM. BAS is supported by core funds from UKRI-NERC to the BEA Team. We deeply appreciate the editorial work of the Associate Editor and the useful comments and suggestions of two reviewers.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aljanabi S. M., Martinez I. (1997). Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 25, 4692–4693. doi: 10.1093/nar/25.22.4692
Allcock A. L., Barratt I., Eléaume M., Linse K., Norman M. D., Smith P. J., et al. (2011). Cryptic speciation and the circumpolarity debate: A case study on endemic Southern Ocean octopuses using the COI barcode of life. Deep-Sea Res. Pt II 58, 242–249. doi: 10.1016/j.dsr2.2010.05.016
Allcock A. L., Strugnell J. M. (2012). Southern Ocean diversity: new paradigms from molecular ecology. Trends Ecol. Evol. 27, 520–528. doi: 10.1016/j.tree.2012.05.009
Amsler C. D., Amsler M. O., Curtis M. D., McClintock J. B., Baker B. J. (2019). Impacts of gastropods on epiphytic microalgae on the brown macroalga Himantothallus grandifolius. Ant Sci. 31, 89–97. doi: 10.1017/S095410219000014
Amsler C. D., Miller L. R., Edwards R. A., Amsler M. O., Engl W., McClintock J. B., et al. (2022). Gastropod assemblages associated with Himantothallus grandifolius, Sarcopeltis Antarctica and other subtidal macroalgae. Ant Sci. 34, 246–255. doi: 10.1017/S0954102022000153
Amsler M. O., Huang Y. M., Engl W., McClintock J. B., Amsler C. D. (2015). Abundance and diversity of gastropods associated with dominant subtidal macroalgae from western Antarctic Peninsula. Polar Biol. 38, 1171–1181. doi: 10.1007/s00300-015-1681-4
Arnaud P. M., Bandel K. (1979). Comments on six species of marine Antarctic Littorinacea (Mollusca, Gastropoda). Tethys 8, 213–230.
Aronson R. B., Thatje S., Clarke A., Peck L. S., Blake D. B., Wilga C. D., et al. (2007). Climate change and invasibility of the Antarctic benthos. Ann. Rev. Ecol. Evol. Syst. 38, 129–154. doi: 10.1146/annurev.ecolsys.38.091206.095525
Baird H. P., Miller K. J., Stark J. S. (2011). Evidence of hidden biodiversity, ongoing speciation and diverse patterns of genetic structure in giant Antarctic amphipods. Mol. Ecol. 20, 3439–3454. doi: 10.1111/j.1365-294X.2011.05173.x
Baird H. P., Shin S., Oberprieler R. G., Hullé M., Vernon P., Moon K. L., et al. (2021). Fifty million years of beetle evolution along the Antarctic Polar Front. Proc. Natl. Acad. Sci. U.S.A. 118, e2017383118. doi: 10.1073/pnas.2017384118
Barker P. F., Diekmann B., Escutia C. (2007). Onset of cenozoic antarctic glaciation. Deep-Sea Res. Pt II 54, 2293–2307. doi: 10.1016/j.dsr2.2007.07.027
Barnes D. K. A., Hodgson D. A., Convey P., Allen C. S., Clarke A. (2006). Incursion and excursion of Antarctic biota: past, present and future. Global Ecol. Biogeogr. 15, 121–142. doi: 10.1111/j.1466-822x.2006.00216.x
Blakeslee A. M. H., Whitman Miller A., Ruiz G. M., Johannesson K., André C., Panova M. (2021). Population structure and phylogeography of two North Atlantic Littorina species with contrasting larval development. Mar. Biol. 168, 117. doi: 10.1007/s00227-021-03918-8
Brey T., Dahm C., Gorny M., Klages M., Stiller M., Arntz W. E. (1996). Do Antarctic benthic invertebrates show an extended level of eurybathy? Ant Sci. 8, 3–6.
Cakil Z. V., Garlasché G., Iakovenko N., Di Cesare A., Eckert E. M., Guidetti R., et al. (2021). Comparative phylogeography reveals consistently shallow genetic diversity in a mitochondrial marker in Antarctic bdelloid rotifers. J. Biogeogr. 48, 1797–1809. doi: 10.1111/jbi.14116
Cantera J. R., Arnaud P. M. (1984). Les gastéropodes prosobranches des Iles Kerguelen et Crozet (Sud de L’Océan Indien) comparaison écologique et particularités biologiques. CNFRA [Comité Natl. Francais Recherches Antarctiques] 56, 1–170.
Chenuil A., Saucède T., Hemery L. G., Eléaume M., Féral J.-P., Améziane N., et al. (2018). Understanding processes at the origin of species flocks with focus on the marine Antarctic fauna. Biol. Rev. 93, 481–504. doi: 10.1111/brv.12354
Clarke A., Crame J. A. (1989). The origin of the Southern Ocean marine fauna. Geol. Soc London 47, 253–268. doi: 10.1144/GSL.SP.1989.047.01.19
Clarke A., Crame J. (1992). The Southern-Ocean benthic fauna and climate change – a historical-perspective. Philos. T R. Soc B 338, 299–309. doi: 10.1098/rstb.1992.0150
Clarke A., Crame J. (2010). Evolutionary dynamics at high latitudes: speciation and extinction in polar marine faunas. Phil Trans. R. Soc B 365, 3655–3666. doi: 10.1098/rstb.2010.0270
Convey P., Stevens M. I., Hodgson D. A., Smellie J. L., Hillenbrand C.-D., Barnes D. K. A., et al. (2009). Exploring biological constraints on the glacial history of Antarctica. Quat. Sci. Rev. 28, 3035–3048. doi: 10.1016/j.quascirev.2009.08.015
Crame J. (2018). Key stages in the evolution of the Antarctic marine fauna. J. Biogeogr. 45, 986–994. doi: 10.1111/jbi.13208
Cumming R. A., Nikula R., Spencer H. G., Waters J. M. (2014). Transoceanic genetic similarities of kelp-associated sea slug populations: long-distance dispersal via rafting? J. Biogeogr. 41, 2357–2370. doi: 10.1111/jbi.12376
Dambach J., Thatje S., Rödder D., Basher Z., Raupach M. J. (2012). Effects of Late-Cenozoic glaciation on habitat availability in Antarctic benthic shrimps (Crustace: Decapoda: Caridae). PloS One 7, e46283. doi: 10.1371/journal.pone.0036283
Damerau M., Marschiner M., Salzburger W., Hanel R. (2012). Comparative population genetics of seven nototothenioid fish species reveals high levels of gene flow along ocean currents in the southern Scotia Arc, Antarctica. Polar Biol. 35, 1073–1086. doi: 10.1007/s00300-012-1155-x
Damerau M., Salzburger W., Hanel R. (2014). Population genetic structure of Lepidonotothen larseni revisited: cyb and microsatellites suggest limited connectivity in the Southern Ocean. Mar. Ecol. Prog. Ser. 517, 251–263. doi: 10.3354/meps11061
Díaz A., Féral J.-P., David B., Saucède T., Poulin E. (2011). Evolutionary pathways among shallow and deep-sea echinoids of the genus Sterechinus in the Southern Ocean. Deep Sea Res. Pt II 58, 205–211. doi: 10.1016/j.dsr2.2010.10.012
Díaz A., Gérard K., González-Wevar C., Maturana C., Féral J.-P., David B., et al. (2018). Genetic structure and demographic inference of the regular sea urchin Sterechinus neumayeri (Meissne 1900) in the Southern Ocean: The role of the last glaciation. PloS One 13, e0197611. doi: 10.1371/journal.pone.0197611
Doellman M. M., Trussel G. C., Grahame J. W., Vollmer S. V. (2011). Phylogeographic analysis reveals a deep lineage split within North Atlantic Littorina saxatilis. Proc. R. Soc B 278, 3175–3183. doi: 10.1098/rspb.2011.0346
Dornburg A., Eytan R. I., Federman S., Pennington J. N., Stewart A. L., Jones C. D., et al. (2016). Molecular data support the existence of two species of the Antarctic fish genus Cryodraco (Channicthyidae). Polar Biol. 39, 1369–1379. doi: 10.1007/s00300-015-1859-9
Drummond A. J., Rambaut A. (2007). BEAST: bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. doi: 10.1186/1471-2148-7-214
Drummond A. J., Suchard M. A., Xie D., Rambaut A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. doi: 10.1093/molbev/mss075
Dueñas L. F., Tracey D. M., Crawford A. J., Wilke T., Alderslade P., Sánchez J. A. (2016). The Antarctic Circumpolar Current as a diversification trigger for deep-sea octocorals. BMC Evol. Biol. 16, 2. doi: 10.1186/s12862-015-0574-z
Dupanloup I., Schneider S., Excoffier L. (2002). A simulated annealing approach to define the genetic structure of populations. Mol. Ecol. 11, 2571–2581. doi: 10.1046/j.1365-294x.2002.01650.x
Edgar R. C. (2004). MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 5, 113. doi: 10.1186/1471-2105-5-113
Excoffier L., Laval G., Schneider S. (2005). Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. Online 1, 37–50. doi: 10.1177/117693430500100003
Fassio G., Modica M. V., Alvaro M. C., Buge B., Salvi D., Oliverio M., et al. (2019). An Antarctic flock under the Thorson’s rule: Diversity and larval development of Antarctic Velutinidae (Mollusca: Gastropoda). Mol. Phylogenet. Evol. 132, 1–13. doi: 10.1016/j.ympev.2018.11.017
Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biotech 3, 294–299.
Forster P., Torroni A., Renfrew C., Röhl A. (2001). Phylogenetic star contraction applied to Asian and Papuan mtDNA evolution. Mol. Biol. Evol. 18, 1864–1881. doi: 10.1093/oxfordjournals.molbev.a003728
Fraser C. I., Nikula R., Ruzzante D. E., Waters J. M. (2012). Poleward bound: biological impacts of Southern Hemisphere glaciation. Trends Ecol. Evol. 27, 462–471. doi: 10.1016/j.tree.2012.04.011
Fraser C. I., Nikula R., Spencer H. G., Waters J. M. (2009). Kelp genes reveal effects of subantarctic sea during the Last Glacial Maximum. Proc. Natl. Acad. Sci. U.S.A. 106, 3249–3253. doi: 10.1073/pnas.0810635106
Fraser C. I., Zuccarello G. C., Spencer H. G., Salvatore L. C., Garcia G. R., Waters J. M., et al (2013). Genetic affinities between trans-oceanic populations of non-buoyant macroalgae in the high latitudes of the southern hemisphere. PLoS ONE 8, e69138. doi:10.1371/journal.pone.0069138
Fraser C. I., Terauds A., Smellie J., Convey P., Chown S. L. (2014). Geothermal activity helps life survive glacial cycles. Proc. Natl. Acad. Sci. U.S.A. 111, 5634–5639. doi: 10.1073/pnas.1321437111
Frugone M. J., López M. E., Segovia N. I., Cole T. L., Lowther A., Pistorius P., et al. (2019). More than the eye can see: Genomic insights into the drivers of genetic differentiation in Royal/Macaroni penguins across the Southern Ocean. Mol. Phylogenet. Evol. 139, 106563. doi: 10.1016/j.ympev.2019.106563
González-Wevar C. A., Hüne M., Cañete J. I., Mansilla A., Nakano T., Poulin E. (2012). Towards a model of postglacial biogeography in shallow marine species along the Patagonian province: lessons from the limpet Nacella magellanica (Gmelin 1791). BMC Evol. Biol. 12, 139. doi: 10.1186/1471-2148-12-139
González-Wevar C. A., Nakano T., Cañete J. I., Poulin E. (2011). Concerted genetic, morphological and ecological diversification in Nacella limpets in the Magellanic Province. Mol. Ecol. 20, 1936–1951. doi: 10.1111/j.1365-294X.2011.05065.x
González-Wevar C. A., Saucède T., Morley T., Chown S. L., Poulin E. (2013). Extinction and recolonization of maritime Antarctica in the limpet Nacella concinna (Strebel 1908) during the last glacial cycle: toward a model of Quaternary biogeography in shallow Antarctic invertebrates. Mol. Ecol. 22, 5221–5236. doi: 10.1111/mec.12465
González-Wevar C. A., Hüne M., Segovia N. I., Nakano T., Spencer H. G., Chown S. L., et al (2017). Following the Antarctic Circumpolar Current: patterns and processes in the biogeography of the limpet Nacella(Mollusca: Patellogastropoda) across the Southern Ocean. J. Biogeogr. 44, 861–874. doi: 10.1111/jbi.12908
González-Wevar C. A., Segovia N. I., Rosenfeld S., Maturana C. S., Jeldres V., Pinochet R., et al. (2022). Seven snail species hidden in one: Biogeographic diversity in an apparently widespread periwinkle in the Southern Ocean. J. Biogeogr. 49, 1521–1534. doi: 10.1111/jbi.14453
González-Wevar C. A., Segovia N. I., Rosenfeld S., Noll D., Maturana C. S., Hüne M., et al. (2021). Contrasting biogeographical patterns in Margarella (Gastropoda: Calliostomatidae: Margarellinae) across the Antarctic Polar Front. Mol. Phylogenet. Evol. 156, 107039. doi: 10.1016/j.ympev.2020.107039
González-Wevar C. A., Segovia N. I., Rosenfeld S., Ojeda J., Hüne M., Naretto J., et al. (2018). Unexpected absence of island endemics: Long-distance dispersal in higher latitude sub-Antarctic Siphonaria (Gastropoda: Euthyneura) species. J. Biogeogr. 45, 874–884. doi: 10.1111/jbi.13174
Guillemin M.-L., Dubrasquet H., Reyes J., Valero M. (2018). Comparative phylogeography of six red algae along the Antarctic Peninsula: extreme genetic depletion linked to historical bottlenecks and recent expansion. Polar Biol. 41, 827–837. doi: 10.1007/s00300-017-2244-7
Guillot G., Mortier F., Estoup A. (2005). GENELAND: a computer package for landscape genetics. Mol. Ecol. Notes 5, 712–715. doi: 10.1111/j.1471-8286.2005.01031.x
Halanych K. M., Mahon A. R. (2018). Challenging the dogma concerning biogeographic patterns of Antarctica and the Southern Ocean. Annu. Rev. Ecol. Evol. Syst. 49, 355–378. doi: 10.1146/annurev-ecolsys-121415-032139
Ho S. Y. W., Lanfear R., Bromham L., Phillips M. J., Soubrier J., Rodrigos A. G., et al. (2011). Time-dependent rates of molecular evolution. Mol. Ecol. 20, 3087–3101. doi: 10.1111/j.1365-294X.2011.05178.x
Ho S. Y. W., Phillips M. J., Cooper A., Drummond A. J. (2005). Time dependency of molecular rate estimated and systematic overestimation of recent divergence times. Mol. Biol. Evol. 22, 1561–1568. doi: 10.1093/molbev/msi145
Ho S. Y. W., Shapiro B., Phyllips M. J., Cooper A., Drummond A. J. (2007). Evidence for time dependency of molecular rate estimates. Syst. Biol. 56, 517–522. doi: 10.1080/10635150701435401
Hoffman J. I., Peck L. S., Hillyard G., Zieritz A., Clark M. S. (2010). No evidence for genetic differentiation between Antarctic limpet Nacella concinna morphotypes. Mar. Biol. 157, 765–778. doi: 10.1007/s00227-009-1360-5
Hoffman J. I., Peck L. S., Linse K., Clarke A. (2011). Strong population genetic structure in a broadcast-spawning Antarctic marine invertebrate. J. Hered 102, 55–66. doi: 10.1093/jhered/esq094
Hudson R. R. (2000). A new statistic for detecting genetic differentiation. Genetics 155, 2011–2014. doi: 10.1093/genetics/155.4.2011
Hüne M., González-Wevar C., Poulin E., Mansilla A., Fernández D. A., Barrera-Oro E. (2015). Low levels of genetic divergence between Harpagifer fish species (Perciformes: Notothenioidei) suggests a Quaternary colonization of Patagonia from the Antarctic Peninsula. Polar Biol. 38, 607–617. doi: 10.1007/s00300-014-1623-6
Ihaka R., Gentleman R. (1996). R: A language for data analysis and graphics. J. Comp. Graph Stat. 5, 299–314. doi: 10.2307/1390807
Iken K. (1999). Feeding ecology of the Antarctic herbivorous gastropod Laevilacunaria Antarctica Martens. J. Exp. Mar. Biol. Ecol. 236, 133–148. doi: 10.1016/S0022-0981(98)00199-3
Ingólfson Ó. (2004). “Quaternary glacial and climate history of Antarctica,” in Quaternary glaciations – extent and chronology, part III: South America, Asia, Australia and Antarctica. Eds. Ehlers J., Gibbard P. L. (Development in Quaternary Science. Elsevier ScienceDirect), 3–43.
Krabbe K., Leese F., Mayer C., Tollrian R., Held C. (2010). Cryptic mitochondrial lineages in the widespread pycnogonid Colossendeis megalonyx Hoek 1881 from Antarctic and Subantarctic waters. Polar Biol. 33, 281–292. doi: 10.1007/s00300-009-0703-5
Lau S. C. Y., Wilson N. G., Silva C. N. S., Strugnell J. M. (2020). Detecting glacial refugia in the Southern Ocean. Ecography 43, 1–18. doi: 10.1111/ecog.04951
Levicoy D., Flores K., Rosenfeld S., Cárdenas L. (2021). Phylogeography and genetic diversity of the microbivalve Kidderia subquadrata, reveals new data from West Antarctic Peninsula. Sci. Rep. 11, 5705. doi: 10.1038/s41598-021-85042-7
Librado P., Rozas J. (2009). DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Littlewood D. T. J., Curini-Galletti M., Herniou E. A. (2000). The interrelationships of Proseriata (Platyhelminthes: Seriata) tested with molecules and morphology. Mol. Phylogenet. Evol. 16, 449–466. doi: 10.1006/mpev.2000.0802
Maggs C. A., Castilho R., Foltz D., Henzler C., Jolly M. T., Kelly J., et al. (2008). Evaluating signatures of glacial refugia for North Atlantic benthic marine taxa. Ecology 89, S108–S122. doi: 10.1890/08-0257.1
Marko P. B. (2004). ‘What’s larvae got to do with it?’ Disparate patterns of post-glacial population structure in two benthic marine gastropod with identical dispersal potential. Mol. Ecol. 13, 597–611. doi: 10.1046/j.1365-294X.2004.02096.x
Marko P. B., Hoffman J. M., Emme S. A., McGovern T. M., Keever C. C., Cox N. (2010). The ‘Expansion–Contraction’ model of Pleistocene biogeography: rocky shores suffer a sea change? Mol. Ecol. 19, 146–167. doi: 10.1111/j.1365-294X.2009.04417.x
Martens van E., Pfeffer G. (1886). Die Mollusken von Süd–Georgien nach der Ausbeute der Deutschen Station 1882–83. Jahr Hamb Wiss. Anst 3, 65–135.
Martin A., Miloslavich P., Díaz Y., Ortega I., Klein E., Troncoso J., et al. (2016). Intertidal benthic communities associated with the macroalgae Iridaea cordata and Adenocystis utricularis in King George Island, Antarctica. Polar Biol. 39, 207–220. doi: 10.1007/s00300-015-1773-1
Matschiner M., Hanel R., Salzburger W. (2009). Gene flow by larval dispersal in the Antarctic notothenioid fish Gobionotothen gibberifrons. Mol. Ecol. 18, 2574–2587. doi: 10.1111/j.1365-294X.2009.04220.x
Maturana C. S., Segovia N. I., González-Wevar C. A., Díaz A., Rosenfeld S., Poulin E., et al. (2020). Evidence of strong small-scale population structure in the Antarctic freshwater copepod Boeckella poppei in lakes on Signy Island, South Orkney Islands. Limnol Oceanogr 65, 2024–2040. doi: 10.1002/lno.11435
McCay R. M., Barrett P. J., Levy R. S., Naish T. R., Golledge N. R., Pyne A. (2016). Antarctic Cenozoic climate history from sedimentary records: ANDRILL and beyond. Phil Trans. R. Soc A 374, 20140301. doi: 10.1098/rsta.2014.0301
McGaughran A., Stevens M. I., Hogg I. D., Carapelli A. (2011). Extreme glacial legacies: A synthesis of the Antarctic springtail phylogeographic record. Insects 2, 62–82. doi: 10.3390/insects2020062
Moon K. L., Chown S. L., Fraser C. I. (2017). Reconsidering connectivity in the sub-Antarctic. Biol. Rev. 92, 2164–2181. doi: 10.1111/brv.12327
Narum S. R. (2006). Beyond Bonferroni: Less conservative analyses for conservation genetics. Cons Genet. 7, 783–787. doi: 10.1007/s10592-005-9056-y
Near T. J., Dornburg A., Kuhn K. L., Eastman J. T., Pennington J. N., Patarnello T., et al. (2012). Ancient climate change, antifreeze, and the evolutionary diversification of Antarctic fishes. Proc. Natl. Acad. Sci. U.S.A. 109, 3434–3439. doi: 10.1073/pnas.1115169109
Nikula R., Fraser C. I., Spencer H. G., Waters J. M. (2010). Circumpolar dispersal by rafting in two subantarctic kelp-dwelling crustaceans. Mar. Ecol-Prog Ser. 405, 221–230. doi: 10.3354/meps08523
Nylander J. A. A. (2004). MrModeltest Version 2. Program Distributed by the Author. Evolutionary Biology Centre, Uppsala University, Uppsala.
Parker R. W., Paige K. N., DeVries A. L. (2002). Genetic variation among populations of the Antarctic toothfish: evolutionary insights and implications for conservation. Polar Biol. 25, 256–261. doi: 10.1007/s00300-001-0333-z
Pearse J. S., Mooi R., Lockhart S. J., Brandt A. (2009). “Brooding and species diversity in the Southern Ocean: Selection for brooders or speciation within brooding clades? ” in Smithsonian at the poles: Contributions to International Polar Year science. Eds. Krupnik I., Lang M. A., Miller S. E. Smithsonian Institution Scholarly Press, Washington, DC USA.
Pollard D., DeConto R. M. (2009). Modeling West Antarctic ice sheet growth and collapse through the past five million years. Nature 458, 329–332. doi: 10.1038/nature07809
Pons O., Petit R. J. (1996). Measuring and testing genetic differentiation with ordered Versus unordered alleles. Genetics 144, 1237–1245. doi: 10.1093/genetics/144.3.1237
Powell A. W. B. (1951). Antarctic and subantarctic mollusca: pelecypoda and gastropoda. Discovery Rep. 26, 47–196. doi: 10.5962/bhl.part.16335
Preston H. B. (1916). Descriptions of eight new species of marine Mollusca from the South Shetland Islands. Ann. Mag Nat. Hist 18, 269–272. doi: 10.1080/00222931608693847
Provan J., Bennett K. D. (2008). Phylogeographic insights into cryptic glacial refugia. Trends Ecol. Evol. 23, 564–571. doi: 10.1016/j.tree.2008.06.010
Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A. (2018). Posterior summarization in Bayesian phylogenetics using Tracer. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Raupach M. J., Thatje S., Dambach J., Rehm P., Misof B., Leese F. (2010). Genetic homogeneity and circum-Antarctic distribution of two benthic shrimp species of the Southern Ocean, Chorismus antarcticus and Nematocarcinus lanceopes. Mar. Biol. 157, 1783–1797. doi: 10.1007/s00227-010-1451-3
Reid D. G., Dyal P., Williams S. T. (2012). A global molecular phylogeny of 147 periwinkle species (Gastropoda, Littorininae). Zool. Scripta 41, 125–136. doi: 10.1111/j.1463-6409
Riesgo A., Taboada S., Avila C. (2015). Evolutionary patterns in Antarctic marine invertebrates: An update on molecular studies. Mar. Genomics 23, 1–13. doi: 10.1016/j.margen.2015.07.005
Rogers A. R., Harpending H. C. (1992). Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. 9, 552–569. doi: 10.1093/oxfordjournals.molbev.a04727
Rosenfeld S., Segovia N. I., Maturana C. S., Aldea C., Saucède T., Brickle P., et al. (2023). A revision of the higher latitude periwinkle species Laevilitorina caliginosa sensu lato. Zool. J. Linn. Soc. (in press). doi: 10.1093/zoolinnean/zlad171
Salzburger W., Ewing G. B., Von Haessler A. (2011). The performance of phylogenetic algorithms in estimating haplotype genealogies with migration. Mol. Ecol. 20, 1952–1963. doi: 10.1111/j.1365-294X.2011.05066.x
Schmider-Martínez A., Maturana C. S., Poveda Y., López-Farran Z., Saucède T., Poulin E., et al. (2023). Laevilacunaria (Mollusca: Gastropoda) in the Southern Ocean: a comprehensive occurrence dataset. Biodivers. Data J. 11, e111982.
Silva S. E., Silva I. C., Madeira C., Sallema R., Paulo O. S., Paula J. (2013). Genetic and morphological variation in two littorinid gastropods: evidence for recent population expansions along the East African coast. Biol. J. Linn. Soc 108, 494–508. doi: 10.1111/j.1095-8312.2012.02041.x
Simone L. R. (2018). Convergence with naticids: phenotypic phylogenetic study on some Antarctic littorinoideans, with description of the zerotulid new genus Pseudonatica, and its presence in Brazil (Mollusca, Caenogastropoda). J. Mar. Biol. Ass UK 98, 1365–1381. doi: 10.1017/S002531541700025X
Simpson R. D., Harrington S. A. (1985). Egg masses of three gastropods, Kerguelenella lateralis (Siphonariidae), Laevilitorina caliginosa and Macquariella hamiltoni (Littorinidae), from Macquarie Island (sub-Antarctic). J. Malacol Soc Aust. 7, 17–28. doi: 10.1080/00852988.1985.10673973
Smith E. A. (1879). Mollusca. An account of the petrological, botanical and zoological collection made in Kerguelen´s Island and Rodriguez during the Transit of “Venus” Expedition in the years 1874–75. Philos. Trans. R. Soc Lond. B 168, 167–192.
Sotelo G., Duvetorp M., Costa D., Panova M., Johannesson K., Faria R. (2020). Phylogeographic history of flat periwinkles, Littorina fabalis and L. obtusata. BMC Evol. Biol. 20, 23. doi: 10.1186/s12862-019-1561-6
Suchard M. A., Weiss R. E., Sinsheimer J. S. (2001). Bayesian selection of continuous-time Markov Chain evolutionary models. Mol. Biol. Evol. 18, 1001–1013. doi: 10.1093/oxfordjournals.molbev.a003872
Tamura K., Stecher G., Kumar S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Thatje S., Hillenbrand C.-D., Larter R. (2005). On the origin of Antarctic marine benthic community structure. Trends Ecol. Evol. 20, 534–540. doi: 10.1016/j.tree.2005.07.010
Thatje S., Hillenbrand C.-D., Mackensen A., Larter R. (2008). Life hung by a thread: endurance of Antarctic fauna in glacial periods. Ecology 89, 682–689. doi: 10.1890/07-0498.1
Thornhill D. J., Mahon A. R., Norenburg J. L., Halanych K. (2008). Open-ocean barriers to dispersal: a test case with the Antarctic Polar Front and the ribbon worm Parborlasia corrugatus (Nemertea: Lineidae). Mol. Ecol. 17, 5104–5117. doi: 10.1111/j.1365-294X.2008.03970.x
Vianna J. A., Noll D., Dantas G. P. M., Petry M. V., Barbosa A., González-Acuña D., et al. (2017). Marked phylogeographic structure of Gentoo penguin reveals an ongoing diversification process along the Southern Ocean. Mol. Phylogenet. Evol. 107, 486–498. doi: 10.1016/j.ympev.2016.12.003
Waters J. M. (2008). Driven by the West Wind Drift? A synthesis of southern temperate marine biogeography, with new directions for dispersalism. J. Biogeogr. 35, 417–427. doi: 10.1111/j.1365-2699.2007.01724.x
Williams S. T., Reid D. G. (2004). Speciation and diversity on tropical rocky shores: a global phylogeny of snail of the genus Echinolittorina. Evolution 58, 2227–2251. doi: 10.1111/j.0014-3820.2004.tb01600.x
Williams S. T., Reid D. G., Littlewood D. T. J. (2003). A molecular phylogeny of the Littorininae (Gastropoda: Littorinidae): unequal evolutionary rates, morphological parallelism, and biogeography of the Southern Ocean. Mol. Phylogenet. Evol. 28, 60–86. doi: 10.1016/S1055-7903(03)00038-1
Wilson N. G., Hunter R. L., Lockhart S. J., Halanych K. M. (2007). Multiple lineage and absence of panmixia in the “circumpolar” crinoid Promachocrinus kerguelensis from the Atlantic sector of Antarctica. Mar. Biol. 152, 895–904. doi: 10.1007/s00227-007-0742-9
Wilson N. G., Schrödl M., Halanych K. M. (2009). Ocean barriers and glaciation: evidence for explosive radiation of mitochondrial lineages in the Antarctic sea slug Doris kerguelenensis (Mollusca, Nudibranchia). Mol. Ecol. 18, 965–984. doi: 10.1111/j.1365-294X.2008.04071.x
Xia X., Xie Z. (2001). DAMBE: Software package for data analysis in Molecular Biology and Evolution. J. Hered. 92, 371–373. doi: 10.1093/jhered/92.4.371
Young E. F., Belchier M., Hauser L., Horsburg G. J., Meredith M. P., Murphy E. J., et al. (2015). Oceanography and life history predict contrasting genetic population structure in two Antarctic fish species. Evol. Appl. 8, 486–509. doi: 10.1111/eva.12259
Keywords: maritime Antarctica, sub-Antarctic, South Georgia, Laevilacunaria antarctica, quaternary glaciations, postglacial recolonization, rafting, dispersal potential
Citation: González-Wevar CA, Poveda Y, Segovia NI, Rosenfeld S, Maturana CS, Jeldres V, Schmider-Martínez A, Gérard K, Spencer HG and Poulin E (2024) Both high and low dispersal? Apparently contradictory genetic patterns in the Antarctic littorinid gastropod Laevilacunaria antarctica. Front. Ecol. Evol. 11:1320649. doi: 10.3389/fevo.2023.1320649
Received: 12 October 2023; Accepted: 28 December 2023;
Published: 06 February 2024.
Edited by:
Jasmine Lee, British Antarctic Survey (BAS), United KingdomReviewed by:
Sally Lau, James Cook University, AustraliaKara Layton, University of Aberdeen, United Kingdom
Copyright © 2024 González-Wevar, Poveda, Segovia, Rosenfeld, Maturana, Jeldres, Schmider-Martínez, Gérard, Spencer and Poulin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Claudio A. González-Wevar, claudio.gonzalez@uach.cl
 Claudio A. González-Wevar
Claudio A. González-Wevar Yarleth Poveda1,2,3
Yarleth Poveda1,2,3  Nicolás I. Segovia
Nicolás I. Segovia Sebastián Rosenfeld
Sebastián Rosenfeld Claudia S. Maturana
Claudia S. Maturana Karin Gérard
Karin Gérard Hamish G. Spencer
Hamish G. Spencer Elie Poulin
Elie Poulin