The Multi-Ocean Distribution of the Hadal Amphipod, Hirondellea dubia Dahl, 1959 (Crustacea, Amphipoda)
- 1School of Natural and Environmental Sciences, Newcastle University, Newcastle upon Tyne, United Kingdom
- 2Minderoo-UWA Deep-Sea Research Centre, School of Biological Sciences and Oceans Institute, The University of Western Australia, Perth, WA, Australia
The hadal zone (depths > 6,000 m) comprises isolated features that host high degrees of endemism, with species inhabiting only a single feature or a group of adjacent features. The amphipod, Hirondellea dubia is an abundant scavenger at the hadal depths of the Kermadec, Tonga, and New Hebrides trenches in the Southwest Pacific Ocean, particularly at depths > 9,500 m. However, several records have hinted that H. dubia may not be restricted to the Southwest Pacific nor be exclusively hadal. Here, we present new records of H. dubia recovered by baited landers deployed to five hadal trenches during three expeditions and assess the geographic and vertical distribution of this species. To rule out the possibility of cryptic diversity, morphological taxonomy was paired with two mitochondrial markers (16S and COI) to test for species boundaries among the new and published molecular data using four delineation approaches (ABGD, CD-HIT, GMYC, and bPTP). We found H. dubia to be a single species and not restricted to the Southwest Pacific or hadal depths. This species’ geographic range extends from the Southwest Pacific to the Mariana Trench (Northwest Pacific), the Clarion-Clipperton Zone (Central Pacific), the South Sandwich Trench (Southern), and the Puerto Rico Trench (North Atlantic). This amphipod’s vertical range spans over half of the ocean’s total depth, between 4,700 and 10,817 m. This study presents an extraordinary geographic range extension to a species long considered endemic to a cluster of three Southwest Pacific trenches and shows a pan-oceanic distribution across extremely isolated hadal features.
Introduction
The deepest oceanic zone, the hadal zone, comprises subduction trenches, fracture zones/trench faults, and troughs that extend from depths of 6,000 to ∼11,000 m (Jamieson et al., 2010). These hadal features are often separated by thousands of kilometers and defined by high hydrostatic pressure, near-freezing temperatures, food scarcity, and geologic instability (Jamieson et al., 2010; Stewart and Jamieson, 2018). As isolated habitats with strong environmental selection pressures, hadal features are thought to host a high number of endemic species, whereby a species inhabits a single trench or a group of adjacent trenches (Wolff, 1959; Belyaev, 1989; Lacey et al., 2016). This concept of hadal endemism has been partially derived from scavenging amphipods, which are well-sampled and conspicuous members of the hadal faunal community (Blankenship et al., 2006), particularly abundant at depths > 8,000 m.
Hirondellea Chevreux, 1889 is a speciose and cosmopolitan genus (Lowry and Stoddart, 2010), with five of the 20 species considered bathymetrically limited to hadal depths and each have restricted geographic distributions. The most notable is H. gigas (Birstein and Vinogradov, 1955), which inhabits depths between 6,770 and 10,897 m in the subduction trench cluster of the Northwest Pacific Ocean (Birstein and Vinogradov, 1955; Dahl, 1959; Kamenskaya, 1977; Hessler et al., 1978; Belyaev, 1989; France, 1993; Kobayashi et al., 2012; Eustace et al., 2013; Ritchie et al., 2015; Jażdżewska and Mamos, 2019). Three Hirondellea species are known only to the Peru-Chile Trench, Southeast Pacific Ocean, specifically H. wagneri Kilgallen, 2015 from 6,173 m, H. sonne Kilgallen, 2015 from 7,050 m, and H. thurstoni Kilgallen, 2015 from 6,173 to 8,072 m (Kilgallen, 2015). Hirondellea dubia Dahl, 1959, the fifth hadal hirondellid, is a dominant scavenger at the Kermadec, Tonga, and New Hebrides trenches in the Southwest Pacific Ocean. At depths greater than 8,000 m, H. dubia is abundant with single traps having recovered more than 12,000 individuals (Blankenship et al., 2006; Lacey et al., 2016; Wilson et al., 2018).
Hirondellea dubia is considered restricted to hadal depths (Jamieson et al., 2011; Wilson et al., 2018). However, several records hint that H. dubia may not be entirely hadal, nor be endemic to the Southwest Pacific Ocean. For example, three out of 585 individuals were found at a 4,700 m site in the New Hebrides Trench (Lacey et al., 2018). Elsewhere in the Pacific Ocean, molecular markers indicate that H. dubia is present at 5,469 m deep at the abyssal plains east of the Mariana Trench (Ritchie et al., 2015), some 6,000 km north of the Kermadec and Tonga trenches, and at the abyssal plains of the Clarion-Clipperton Zone (Bribiesca-Contreras et al., 2021). The New Hebrides and Mariana trench observations are anecdotal, as errors in specimen curation could not be unequivocally ruled out. Outside the Pacific Ocean, H. dubia inhabits between 6,640 and 8,265 m in the South Sandwich Trench, which spans between the Southern and South Atlantic oceans (Jamieson et al., 2021b). Yet, this identification was based solely on morphology and deemed sensu lato. As deep-sea amphipod taxonomy is challenged by cryptic diversity (Havermans et al., 2011, 2013; Jazdzewska et al., 2021), the South Sandwich Trench identification needs confirmation through molecular taxonomy.
The records of H. dubia at abyssal depths and the South Sandwich Trench pose questions about whether they represent the distribution of H. dubia or cryptic species. Here, we present new collection data for H. dubia from the global Five Deeps Expedition and two Mariana Trench expeditions and place it in context with historical records of H. dubia. Given the widespread phenomenon of cryptic diversity in the deep sea, we aimed to rule out the possibility that cryptic species that are morphologically indistinguishable from H. dubia exist outside the Southwest Pacific Ocean. We applied a taxonomic feedback loop approach (Page et al., 2005) by pairing morphological taxonomy with two mitochondrial markers, namely 16S ribosomal DNA (16S) and cytochrome c oxidase 1 (COI), to test for species boundaries among the new material and published data from the Pacific Ocean. The findings of this global study begin to test the concept of hadal endemism and expand our understanding of species distributions across the deepest marine zone.
Materials and Methods
Amphipod Recovery and Morphological Identification
Three sampling expeditions recovered scavenging amphipods by free-fall autonomous landers (Jamieson et al., 2009). Two expeditions sampled the Mariana Trench: the “HADES-M” cruise FK141109 in 2014 on the RV Falkor (5,156–10,545 m) and the “FISH2017” cruise SY1615 in 2017 on the TV Shinyo-Maru (8,000–10,890 m). Between 2018 and 2019, the Five Deeps Expedition deployed landers from the DSSV Pressure Drop to the Puerto Rico Trench (750–8,380 m), South Sandwich Trench (6,044–8,266 m), Agulhas Fracture Zone (5,493 m), Java Trench (5,760–7,176 m), Diamantina Fracture Zone (7,009 m), San Cristobal Trench (6,515–8,407 m), Santa Cruz Trench (5,906–8,428 m), Mariana Trench (2,100–10,924 m), Tonga Trench (6,793–10,817), and Molloy Hole (5,547–5,591 m). Details on the deployment sites that recovered H. dubia are in Table 1. See Drazen (2015), Jamieson et al. (2021a), and Jamieson and Linley (2021) for more complete expedition details.
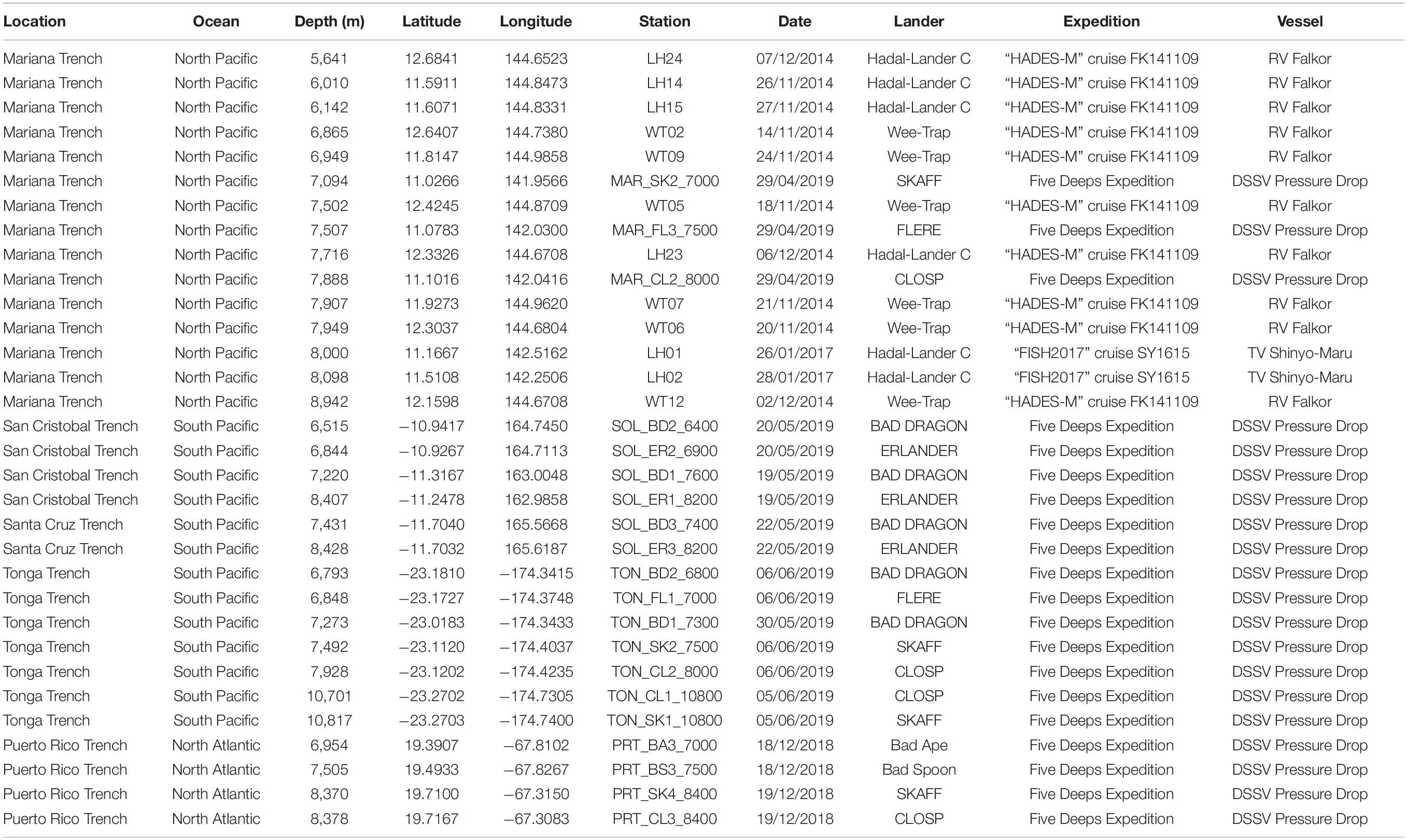
Table 1. Locations and baited lander deployments where the amphipod Hirondellea dubia was recovered during this study.
Each lander was equipped with an invertebrate funnel trap baited with whole mackerel (Scombrus sp.) and a pressure and temperature sensor. Landers stayed at depth for 7–12 h. Pressure (dbar) was converted to depth (m) following Saunders (1981).
Amphipods were preserved in 70% ethanol. Specimens were sorted into morphotypes based on external characters using a stereomicroscope (Wild Heerbrugg M8). We selected a subset of Hirondellea individuals from each location for species-level identification following Dahl (1959), Lowry and Stoddart (2010), and Kilgallen (2015). Appendages were dissected, mounted with glycerol, and imaged with a Leica DMi8 inverted microscope and DFC295 camera.
Historical records for H. dubia are compiled in Supplementary Material 1 (Dahl, 1959; Jamieson et al., 2011, 2021b; Ritchie et al., 2015; Lacey et al., 2016; Leduc and Wilson, 2016; Wilson et al., 2018; Bribiesca-Contreras et al., 2021). Absence records were inferred, with an absence defined as an instance when a published species list for a hadal feature included Lysianassoidea species but not H. dubia (Dahl, 1959; Hessler et al., 1978; Belyaev, 1989; France, 1993; Jamieson, 2015; Lacey et al., 2016; Jażdżewska and Mamos, 2019; Weston et al., 2021b).
DNA Extraction and PCR Amplification
A total of 13 individuals of H. dubia from Mariana, Puerto Rico, South Sandwich, San Cristobal, Santa Cruz, and Tonga trenches were selected for molecular taxonomy. Only 10 individuals were sequenced at both markers. PCR amplification failed for a Tonga Trench and a San Cristobal Trench individual and the Santa Cruz Trench individuals, likely due to poor DNA quality resulting from lander recovery (Dixon et al., 2004). One H. gigas individual from 7,949 m in the Mariana Trench (Table 1) and two Hirondellea sp. individuals from 6,714 to 8,052 m in the Atacama Trench (Weston et al., 2021a) were included in the molecular taxonomy. A total of 24 sequences have been deposited on GenBank (Supplementary Material 2).
Total genomic DNA was extracted with the Bioline ISOLATE II Genomic DNA Kit. The two mitochondrial regions were PCR amplified following Ritchie et al. (2015) with AMPH1 (France and Kocher, 1996) and “Drosophila-type” 16SBr (Palumbi et al., 1991) primers for 16S (260 bp) and LCO1490 and HCO12198 (Folmer et al., 1994) primers for COI (624 bp). PCR products were cleaned with Exonuclease 1 and Antarctic Phosphatase (New England Biolabs) and sequenced on an ABI 3730XL (Eurofins Genomics, Germany). Electropherograms were examined in MEGA X (Kumar et al., 2018), and the primers and ambiguous sequences were manually clipped. COI sequences were translated into amino acids to confirm the absence of stop codons. Sequences were initially compared with species diagnostic barcodes using the BLASTn website1 and the existing Barcode Index Numbers (BINs) on the BOLD v4 website2 (Ratnasingham and Hebert, 2013).
Molecular Dataset Assembly and Species Delimitation
Datasets for each mitochondrial marker were built to assess for cryptic diversity among H. dubia across the Pacific, Southern, and Atlantic oceans. New sequences were supplemented with data that was publicly available by October 2021 on GenBank and the Barcode of Life Data System (BOLD) (Ratnasingham and Hebert, 2007). Published sequences for Alicella gigantea Chevreux, 1899 were chosen for the outgroup (Weston et al., 2020), as this deep-sea scavenging amphipod is in a separate superfamily with enough phylogenetic distance (Ritchie et al., 2015). Alignments were constructed using the MAFFT v7 (Katoh et al., 2019) webserver3 with the FFT-NS-1 strategy and trimmed to equal length. The four alignment datasets were: (1) complete-16S with 36 individuals (176 bp), (2) reduced-16S with 7 haplotypes (176 bp), (3) complete-COI with 27 individuals (500 bp), and (4) reduced-COI with 17 individuals (500 bp). The compiled datasets are presented in Supplementary Material 2.
Four molecular species delimitation methods were applied to test for the number of Molecular Operational Taxonomic Units (MOTUs) or species, with two distance-based and two tree-based methods. The first distance-based method was the Automatic Barcode Gap Discovery (ABGD) (Puillandre et al., 2012), which was conducted on the ABGD webserver4 with the JC69 Jukes-Cantor parameter and default setting. CD-HIT-EST (Li and Godzik, 2006) was the second distance-based method and executed on the CD-HIT Suite webserver5 (Huang et al., 2010) using default settings and a predefined threshold of 0.976 (Kniesz et al., 2022).
The two tree-based phylogenetic delineation approaches were the Generalized Mixed Yule Coalescent (GMYC) likelihood method (Pons et al., 2006; Monaghan et al., 2009) and the Bayesian implementation of the Poisson Tree Processes (bPTP) (Zhang et al., 2013). Both approaches need an input tree, which was inferred using the Bayesian Evolutionary Analysis by Sampling Trees software package v1.10.4 (Suchard et al., 2018). The Hasegawa, Kishino, and Yano model with gamma distribution (Hasegawa et al., 1985) was identified in MEGA X (Kumar et al., 2018) as the optimal evolutionary models for both genes based on the Bayesian information criterion. For GMYC, the ultrametric trees were constructed with the parameters of a normalized exponential relaxed clock and a Yule process of speciation. For the bPTP trees, an uncorrelated relaxed clock was selected. Two independent runs were executed for 40M generations, sampling every 10,000 generations, for each gene and approach. Outputs were reviewed with Tracer v1.7 to ensure convergence (effective sample size > 200) and merged in LogCombiner v1.8.4. The maximum clade credibility tree was determined by TreeAnnotator v1.8.4, after burning the first 10% of states. For GYMC, the number of delimited species was determined through the “gymc” function in the splits v1.0-20 package (Ezard et al., 2021) in R v3.6.3 (R Core Team, 2020). For the bPTP method, the unrooted tree was analyzed on the bPTP webserver6 for 200,000 generations, thinning of 100, and burn-in of 25%.
The Bayesian tree of the reduced-COI dataset is used as the graphical presentation of the species delimitation results and annotated in Inkscape v0.92.2 (The Inkscape Team, 2017).
Results
Presence Records and Morphological Taxonomy
Hirondellea dubia was recovered during 14 baited lander deployments to the Mariana Trench (5,641–8,942 m), four to the San Cristobal Trench (6,515–8,407 m), two to the Santa Cruz Trench (7,431–8,428 m), seven to the Tonga Trench (6,793–10,817 m), and four to the Puerto Rico Trench (6,954–8,378 m) (Table 1 and Figures 1, 2), which adds to the 45 historical records from the Kermadec, Tonga, New Hebrides, Mariana, and South Sandwich trenches and the Clarion-Clipperton Zone (Supplementary Material 1). Records included the deepest points of the Tonga, Puerto Rico, and South Sandwich trenches (Bongiovanni et al., 2021). No hirondellids were recovered from the Java Trench and Diamantina Fracture Zone, nor in the near-hadal depths of the Agulhas Fracture Zone and Molloy Hole.
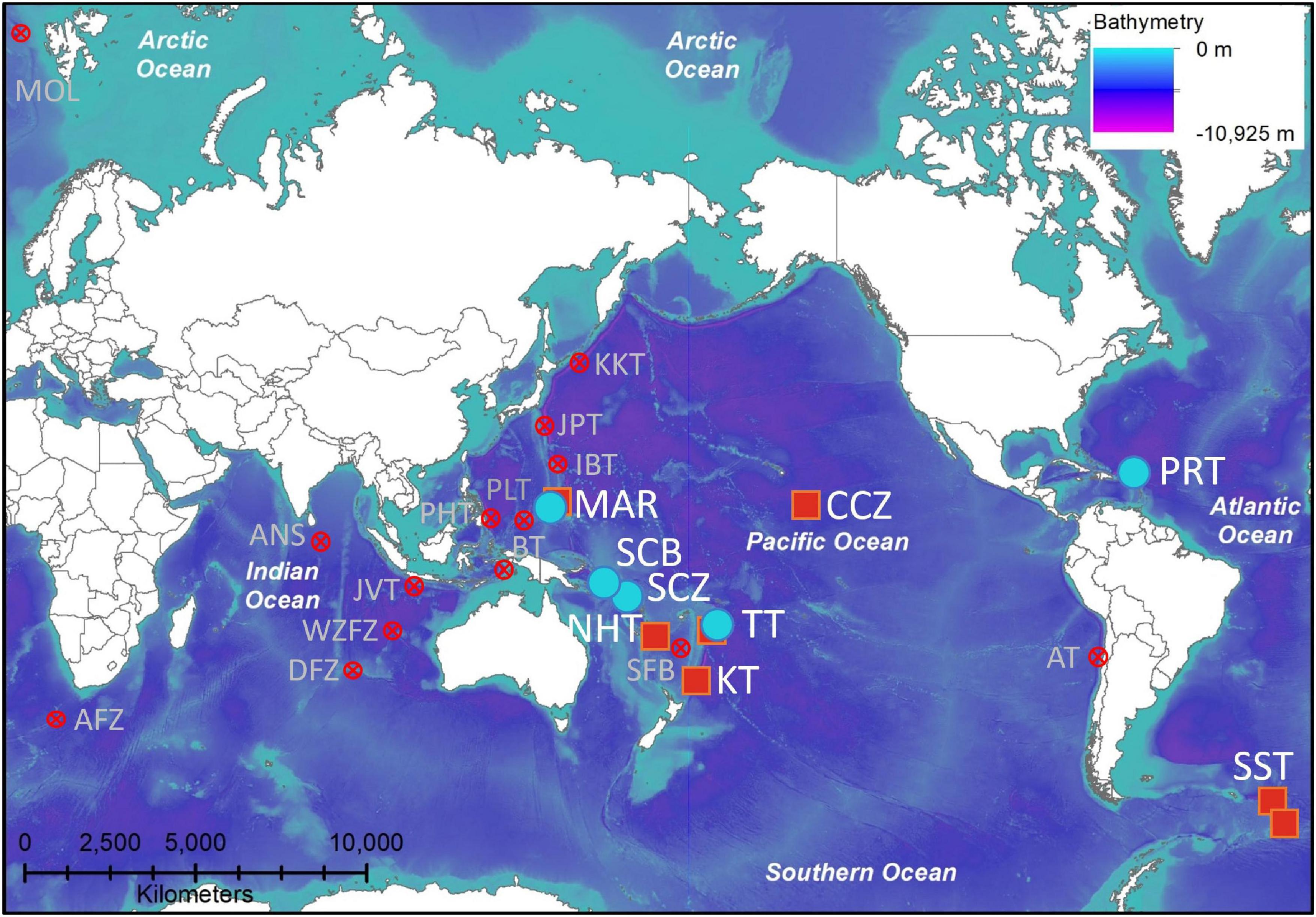
Figure 1. Geographic distribution of the amphipod Hirondellea dubia, with new records (blue circle) compiled with historical records (red square). Features with depths > 4,700 m where H. dubia could be expected to be present but has not been found (red cross). Locations are abbreviated as, AFZ, Agulhas Fracture Zone; ANS, Afanasy-Nikitin Seamount; AT, Atacama Trench; BT, Banda Trench; CCZ, Clarion-Clipperton Zone; DFZ, Diamantina Fracture Zone; IBT, Izu-Bonin Trench; JAV, Java Trench; JPT, Japan Trench; KT, Kermadec Trench; MAR, Mariana Trench; MOL, Molloy Hole; NHT, New Hebrides Trench; PHT, Philippine Trench; PLT, Palau Trench; PRT, Puerto Rico Trench; SBF, South Fiji Basin (only 4,100 m); SST, South Sandwich Trench; SCB, San Cristobal Trench; SCZ, Santa Cruz Trench; TT, Tonga Trench; WZFZ, Wallaby-Zenith Fracture Zone.
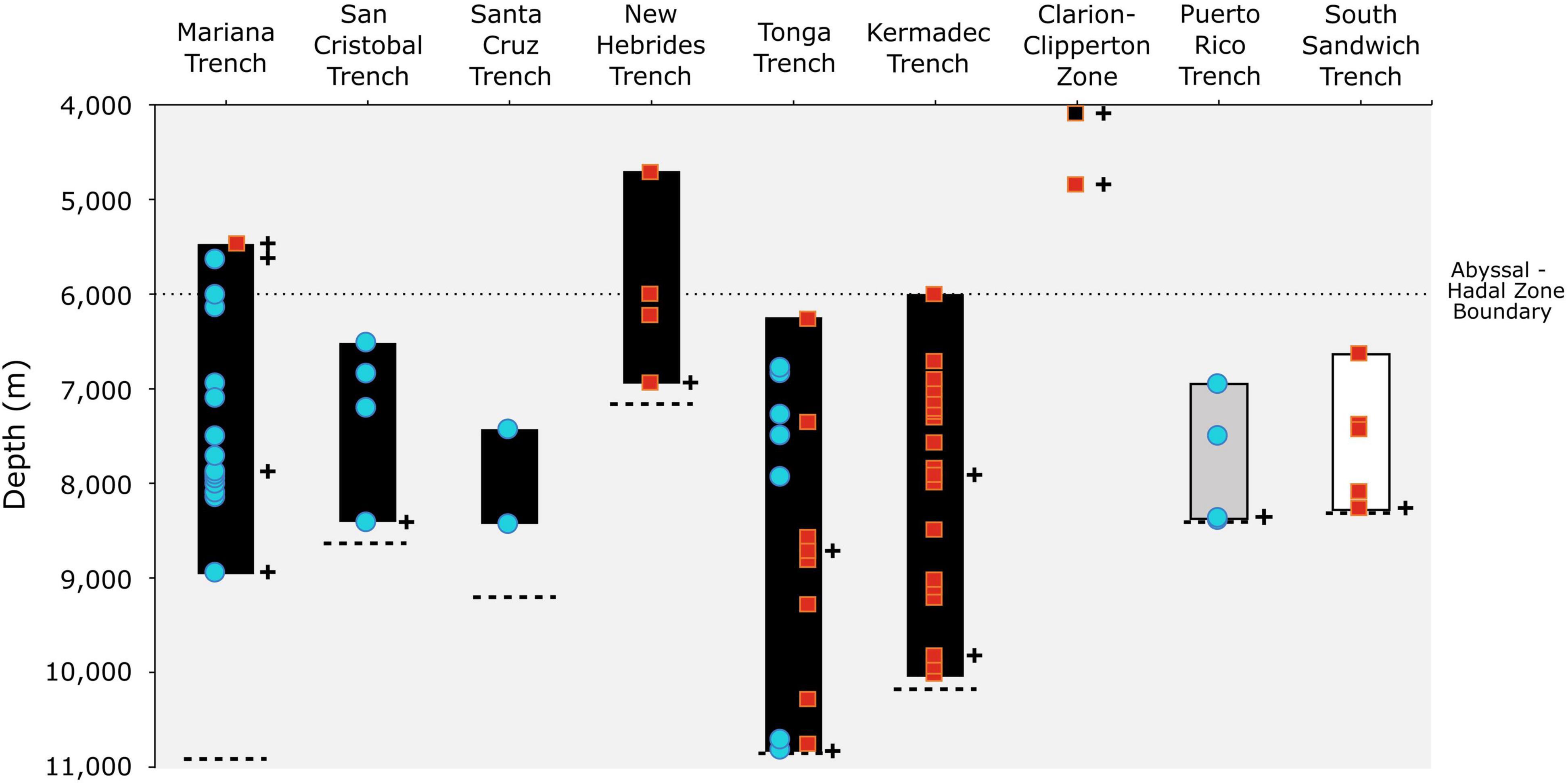
Figure 2. Vertical distribution of the amphipod Hirondellea dubia (solid bar) across nine features in the Pacific (black), North Atlantic (gray), and Southern/South Atlantic (white) oceans with new records (blue circle) and historical records (red square). Cross symbols indicate records with corresponding molecular data, namely 16S and COI. The black square at the Clarion-Clipperton Zone represents the molecular record that was not delimited as H. dubia. The dashed lines represent the maximum trench depth (Stewart and Jamieson, 2018; Bongiovanni et al., 2021).
Examined specimens were morphologically consistent with the original taxonomic species description of H. dubia (Dahl, 1959). In particular, the maxilla 2 outer plate had an 8/3 crown setal-tooth arrangement, the maxilliped inner plate had a concave apical margin with 1 small plumose seta and 1 larger slender seta, the gnathopod 1 dactyl lacked subterminal spines on the inner margin, the gnathopod 2 palm was minutely subchelate, the uropod 2 inner ramus were not incised, the uropod 3 rami were subequal, the telson cleft was one-quarter of its length, and two setae were present at the tips of the telson lobes. No apparent phenotypic variation was observed among individuals from different features.
Species Delimitation
Overall, the four species delimitation methods identified one MOTU for H. dubia based on the 16S and COI markers and separated it from H. gigas and Hirondellea sp. AT-2018 (Figure 3). Additionally, the H. dubia COI sequences were matched to the existing BIN BOLD: AAX1198 and H. gigas to BOLD: ADD2499. Hirondellea sp. AT-2018 was not matched to an existing BIN. Amphipods previously identified and published as Uristes sp. nov. from the Tonga Trench (Blankenship and Levin, 2007; Ritchie et al., 2015) were placed within the H. dubia clade. From the Clarion-Clipperton Zone, the two individuals from 4,865 m were placed in the H. dubia clade. However, the amphipod from 4,084 m, while published on GenBank as H. dubia (Bribiesca-Contreras et al., 2021), was placed in a separate genetic lineage and not matched to the existing H. dubia BIN, BOLD: AAX1198. Record of that individual does not exist in the APEI 1 species list (Bribiesca-Contreras et al., 2021), thus likely more indicative of a data curation error or misidentification than a true presence (Supplementary Material 2).
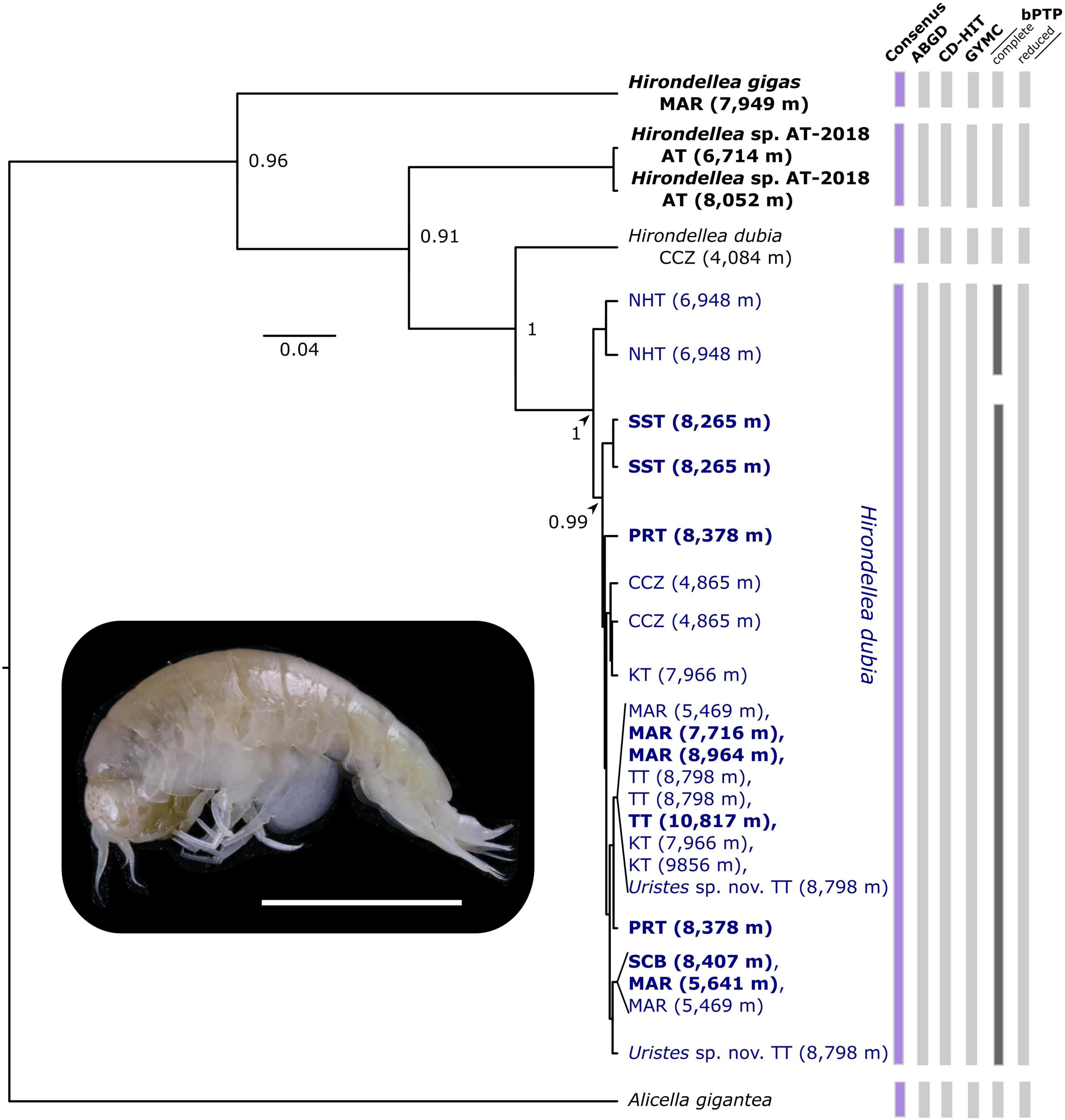
Figure 3. Bayesian inferred tree showing species boundaries among the amphipod Hirondellea dubia (blue) individuals based on COI. Study-generated sequences are in bold. Results from the four molecular species delimitation methods are shown with vertical bars, with disagreements (dark gray) and consensus (purple) highlighted. Branch nodes show posterior probabilities > 0.7. Locations are abbreviated as, AT, Atacama Trench; CCZ, Clarion-Clipperton Zone; KT, Kermadec Trench; MAR, Mariana Trench; NHT, New Hebrides Trench; PRT, Puerto Rico Trench; SST, South Sandwich Trench; SCB, San Cristobal Trench; TT, Tonga Trench. Tip labels show collected depth (meters). See Supplementary Material 2 for the GenBank accession numbers. Photograph of H. dubia from 8,379 m at the Puerto Rico Trench, Atlantic Ocean (scale 5 mm).
Minor disagreements were present among the bPTP approaches (Figure 3), resolving a different number of MOTUs, with 5 in the complete-16S dataset, 4 in the reduced-16S dataset, 6 in the complete-COI dataset, and 5 in the reduced-COI dataset. Oversplitting occurred within the H. dubia clade, where the Mariana, New Hebrides, and San Cristobal trench individuals were a separate MOTU in the complete-16S dataset, and the New Hebrides Trench amphipods were a distinct MOTU in the complete-COI dataset.
Discussion
The hadal zone comprises isolated features that host high levels of species unique to a single feature or a group of adjacent features. Conversely, we found that H. dubia is an amphipod that challenges the concept of hadal endemism. Our combined morphology taxonomy and species delimitation with 16S and COI molecular markers reveal that H. dubia is not restricted to a Southwest Pacific trench cluster but has a multi-oceanic distribution across the Pacific, Southern, and Atlantic oceans, and a vertical range, spanning 6,117 m.
Increasing common in the deep sea, species with wide distribution often turn out to be composed of cryptic species discernible by molecular taxonomy (Bickford et al., 2007; Glazier and Etter, 2014; Brasier et al., 2016). One iconic example is the scavenging amphipod Eurythenes gryllus (Lichtenstein in Mandt, 1822), which was considered to inhabit every ocean between 550 and ∼8,000 m but integrative taxonomy revealed this to be a species complex of at least 12 species (Havermans et al., 2013; Havermans, 2016). Considering this trend, we expected H. dubia to be a species complex, with morphological differences and distinct genetic lineages. However, our dual lines of evidence provided congruent data to strongly suggest that H. dubia is a single species (Figure 3). This study expands its known distribution to the Puerto Rico, Santa Cruz, and San Cristobal trenches, confirms its presence at the Mariana and South Sandwich trenches, and verifies that the historical records of H. dubia and Uristes sp. nov. at the Kermadec, Tonga, and New Hebrides trenches, and the Clarion-Clipperton Zone, except for the record at 4,084 m, are all H. dubia (Figure 1).
Among hadal taxa, H. dubia is an outlier with a multi-oceanic distribution. For example, ∼70% of hadal amphipods, 65% of hadal isopods, and nearly all hadal snailfishes are known to single hadal feature (Belyaev, 1989; Elsner et al., 2015; Jamieson, 2015; Jamieson et al., 2021a). A similar cosmopolitan hadal amphipod is Bathycallisoma schellenbergi (Birstein and Vinogradov, 1958) present at 10 hadal features across the Pacific, Atlantic, Indian, and Southern oceans (Kilgallen and Lowry, 2015; Jamieson et al., 2021b; Weston et al., 2021b). Their wide distributions lead to questions about the degree of connectivity and population isolation. Hirondellea dubia could be a strong taxon to investigate the extent to which hadal features function as independent units by applying high-throughput sequencing methods (Taylor and Roterman, 2017), as the abyssal depth records, albeit very few, indicate that populations may not be restricted to subduction trenches, and thus a possibility of dispersal between trenches.
Hirondellea dubia has a vertical range that covers over half the ocean’s total depth, between 4,700 and 10,817 m. A eurybathic range is not uncommon for deep-sea amphipods, such as ∼4,000 m for H. gigas, ∼3,500 m for Eurythenes maldoror and Eurythenes magellanicus (Havermans, 2016), and ∼6,000 m for A. gigantea (Jamieson et al., 2013). Curiously, the vertical distribution of H. dubia varies among the trenches, with the Mariana Trench appearing as an outlier (Figure 2). At the other seven trenches, H. dubia was found either at or within ∼700 m of the deepest point, which is likely due to sampling design than true absence, particularly for the Santa Cruz Trench. Whereas H. dubia was absent from the deepest ∼2,000 m of the Mariana Trench, despite the next two deployments being at 8,964 and 9,059 m, thus ruling out sampling bias. Another explanation may be that H. dubia is not biochemically capable to survive the deepest ∼2,000 m of the Mariana Trench (Downing et al., 2018). Yet, that is refuted given that H. dubia is present at Horizon Deep, the deepest point of Tonga Trench and a mere 107 m shallower than Challenger Deep (Bongiovanni et al., 2021). An alternative hypothesis may be related to food distribution across the trench axes and ecological interactions between scavenging species. Resource competition has been proposed in the Kermadec and New Hebrides trenches, with H. dubia largely absent from the depths that corresponded to the maximum densities of the larger B. schellenbergi (Lacey et al., 2018). While testing these hypotheses is outside the study’s scope, the distribution of H. dubia across trenches indicates that the drivers of vertical distribution are more complex than solely tolerance to hydrostatic pressure.
Contrary to the geographic range extension, H. dubia was absent at hadal and near-hadal depths in the Arctic, Indian, and the South Atlantic oceans (Figure 1). It is unclear if this is a true absence or sampling bias. Absence from the Agulhas Fracture Zone and Molloy Hole could be due to their abyssal depths (Bongiovanni et al., 2021; Jamieson and Linley, 2021). Whereas the Diamantina Fracture Zone is hadal and at the same latitude as the Kermadec Trench. Furthermore, the Java Trench is also hadal and much closer to the western Pacific Ocean trenches than the South Sandwich or Puerto Rico trenches (Stewart and Jamieson, 2018). These absences combined with the known Wallaby-Zenith Fracture Zone absence (Weston et al., 2021b), suggest that H. dubia does not inhabit the Indian Ocean. This absence cannot be readily explained by other known environmental factors, such as temperature, as the Indian Ocean bottom temperatures lie between the warmer Mariana Trench and colder South Sandwich Trench (Jamieson et al., 2021a). Therefore, an explanation for the absence of H. dubia in the Indian Ocean stays obscure.
In conclusion, H. dubia is a single species with an extensive geographic and vertical range. This species exemplifies the gaps in our understanding of species distribution across the hadal zone. Moreover, the wide distribution of H. dubia raises questions regarding deep ocean biogeography and speciation. For example, what is the degree of connectivity among feature populations? Further, how did H. dubia spread to such distant locations such as the Puerto Rico and South Sandwich trenches but not the Atacama Trench? These questions should begin being resolved as hadal research expands more comprehensively across multiple, trans-oceanic hadal features. Hirondellea dubia represents an important hadal species from which to test eco-evolutionary questions, and expeditions on the scale of the Five Deeps Expedition will be paramount in recovering the specimens to answer these questions.
Data Availability Statement
Physical samples are housed at The University of Western Australia. Sequences were deposited in GenBank. Hirondellea dubia sequences can be accessed under accession numbers OK641752–OK641759 and OK641761 for 16S and OK639157–OK639165 for COI. Hirondellea gigas sequences can be accessed under accession numbers OK641761 for 16S and OK639181 for COI. Hirondellea sp. AT-2018 sequences can be accessed under accession numbers OL376314–OL376314 for 16S and OL379343–OL379344 for COI. See Supplementary Material 2 for specifics.
Author Contributions
AJ: conceptualization and funding acquisition. JW: investigation. AJ and JW: resources, writing – original draft, writing, review, and editing. Both authors contributed to the article and approved the submitted version.
Funding
Sea time and logistics for the Five Deeps Expedition were supported and funded by Victor Vescovo of Caladan Oceanic LLC (United States). The “Hades-M” expedition was funded by the Schmidt Ocean Institute. DNA barcoding was funded by the “Hadal Zones of Our Overseas Territories” by the Darwin Initiative funded by the UK Government (DPLUS093). Expedition participation of AJ was supported by Newcastle University’s Research Infrastructure Fund (RiF), Exploration of Extreme Ocean Environments, awarded to AJ.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the captain and crew of the following expeditions: RV Falkor FK141109 “Hades-M,” TV Shinyo Maru SY1615 “Fish2017,” and the DSSV Pressure Drop Five Deeps Expedition. We extend thanks to Victor Vescovo and Cassie Bongiovanni (Caladan Oceanic LLC, United States), Patrick Lahey (Triton Submarines LLC, United States), Rob McCallum (EYOS Expeditions, United Kingdom), Hiroshi Kitazato (Tokyo University of Marine Science and Technology, Japan), Heather Stewart (British Geological Survey, United Kingdom), Thomas Linley (Armatus Oceanic, United Kingdom), Carolina González (Universidad de Concepción, Chile), and Leah Brown, Michael Stamford-Bewlay, and Jennifer Wainwright (Newcastle University, United Kingdom). We are grateful for the constructive reviews from the reviewers and editor to help focus this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.824640/full#supplementary-material
Footnotes
- ^ https://blast.ncbi.nlm.nih.gov/
- ^ https://www.boldsystems.org/index.php/IDS_OpenIdEngine
- ^ https://mafft.cbrc.jp/alignment/server/
- ^ https://bioinfo.mnhn.fr/abi/public/abgd/abgdweb.html
- ^ http://weizhong-lab.ucsd.edu/cdhit_suite/cgi-bin/index.cgi?cmd=Server%20home
- ^ https://species.h-its.org/
References
Bickford, D., Lohman, D. J., Sodhi, N. S., Ng, P. K. L., Meier, R., Winker, K., et al. (2007). Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 22, 148–155. doi: 10.1016/J.TREE.2006.11.004
Birstein, J. A., and Vinogradov, M. E. (1955). Pelagicheskie gammaridy (Amphipoda, Gammaridea) Kurilo-Kamchatskoi Vpadiny [Pelagic gammarids (Amphipoda, Gammaridea) at the Kourilo-Kamtchatka Trench]. Tr. Inst. Okeanol. Akad. Nauk S.S.S.R. 12, 210–287.
Birstein, J. A., and Vinogradov, M. E. (1958). Pelagicheskie gammaridy (Amphipoda, Gammaridea) severo-zapadnoi chasti Tikhogo Okeana. [Pelagic Gammaridea from the northwestern Pacific Ocean]. Tr. Inst. Okeanol. Akad. Nauk S.S.S.R. 27, 219–257.
Blankenship, L. E., and Levin, L. A. (2007). Extreme food webs: foraging strategies and diets of scavenging amphipods from the ocean’s deepest 5 kilometers. Limnol. Oceanogr. 52, 1685–1697. doi: 10.4319/lo.2007.52.4.1685
Blankenship, L. E., Yayanos, A. A., Cadien, D. B., and Levin, L. A. (2006). Vertical zonation patterns of scavenging amphipods from the hadal zone of the Tonga and Kermadec trenches. Deep Sea Res. Part I Oceanogr. Res. Pap. 53, 48–61. doi: 10.1016/j.dsr.2005.09.006
Bongiovanni, C., Stewart, H. A., and Jamieson, A. J. (2021). High-resolution multibeam sonar bathymetry of the deepest place in each ocean. Geosci. Data J. 0:1–16. doi: 10.1002/gdj3.122
Brasier, M. J., Wiklund, H., Neal, L., Jeffreys, R., Linse, K., Ruhl, H., et al. (2016). DNA barcoding uncovers cryptic diversity in 50% of deep-sea Antarctic polychaetes. R. Soc. Open Sci. 3:160432. doi: 10.1098/RSOS.160432
Bribiesca-Contreras, G., Dahlgren, T. G., Horton, T., Drazen, J. C., Drennan, R., Jones, D. O. B., et al. (2021). Biogeography and connectivity across habitat types and geographical scales in Pacific abyssal scavenging amphipods. Front. Mar. Sci. 8:1028. doi: 10.3389/FMARS.2021.705237
Dixon, D. R., Pruski, A. M., and Dixon, L. R. J. (2004). The effects of hydrostatic pressure change on DNA integrity in the hydrothermal-vent mussel Bathymodiolus azoricus: implications for future deep-sea mutagenicity studies. Mutat. Res. 552, 235–246. doi: 10.1016/J.MRFMMM.2004.06.026
Downing, A. B., Wallace, G. T., and Yancey, P. H. (2018). Organic osmolytes of amphipods from littoral to hadal zones: increases with depth in trimethylamine N-oxide, scyllo-inositol and other potential pressure counteractants. Deep Sea Res. Part I Oceanogr. Res. Pap. 138, 1–10. doi: 10.1016/j.dsr.2018.05.008
Drazen, J. (2015). Cruise Report-FK141109 Studying the Biology and Geology of the Mariana Trench: The Deepest Place on Earth (aka HADES-M). Available online at: https://datadocs.bco-dmo.org/d3/data_docs/Mariana_Perspectives/FK141109_Cruise_Report_JDC_2015-01-12.pdf (accessed January 11, 2022)
Elsner, N. O., Malyutina, M. V., Golovan, O. A., Brenke, N., Riehl, T., and Brandt, A. (2015). Deep down: isopod biodiversity of the Kuril-Kamchatka abyssal area including a comparison with data of previous expeditions of the RV Vityaz. Deep Sea Rese. Part II Top. Stud. Oceanogr. 111, 210–219. doi: 10.1016/j.dsr2.2014.08.007
Eustace, R. M., Kilgallen, N. M., Lacey, N. C., and Jamieson, A. J. (2013). Population structure of the hadal amphipod Hirondellea gigas (Amphipoda: Lysianassoidea) from the Izu-Bonin Trench. J. Crustac. Biol. 33, 793–801. doi: 10.1163/1937240X-00002193
Ezard, T., Fujisawa, T., and Barraclough, T. (2021). Splits: SPecies’ LImits by Threshold Statistics. Available online at: https://R-Forge.R-project.org/projects/splits/ (accessed January 4, 2022).
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299.
France, S. C. (1993). Geographic variation among three isolated population of the hadal amphipod Hirondellea gigas. (Crustacea: Amphipoda: Lysianassoidea). Mar. Ecol. Prog. Ser. 92, 277–287. doi: 10.1007/s10528-019-09935-z
France, S. C., and Kocher, T. D. (1996). Geographic and bathymetric patterns of mitochondrial 16s rRNA sequence divergence among deep-sea amphipods, Eurythenes gryllus. Mar. Biol. 126, 633–643. doi: 10.1007/bf00351330
Glazier, A. E., and Etter, R. J. (2014). Cryptic speciation along a bathymetric gradient. Biol. J. Linn. Soc. 113, 897–913. doi: 10.1111/BIJ.12389
Hasegawa, M., Kishino, H., and Yano, T. (1985). Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22, 160–174. doi: 10.1007/BF02101694
Havermans, C. (2016). Have we so far only seen the tip of the iceberg? exploring species diversity and distribution of the giant amphipod Eurythenes. Biodiversity 17, 12–25. doi: 10.1080/14888386.2016.1172257
Havermans, C., Nagy, Z. T., Sonet, G., de Broyer, C., and Martin, P. (2011). DNA barcoding reveals new insights into the diversity of Antarctic species of Orchomene sensu lato (Crustacea: Amphipoda: Lysianassoidea). Deep Sea Res. Part II Top. Stud. Oceanogr. 58, 230–241. doi: 10.1016/J.DSR2.2010.09.028
Havermans, C., Sonet, G., d’Udekem d’Acoz, C., Nagy, Z. T., Martin, P., Brix, S., et al. (2013). Genetic and morphological divergences in the cosmopolitan deep-sea amphipod Eurythenes gryllus reveal a diverse abyss and a bipolar species. PLoS One 8:e74218. doi: 10.1371/journal.pone.0074218
Hessler, R. R., Ingram, C. L., Aristides Yayanos, A., and Burnett, B. R. (1978). Scavenging amphipods from the floor of the Philippine Trench. Deep Sea Res. 25, 1029–1047. doi: 10.1016/0146-6291(78)90585-4
Huang, Y., Niu, B., Gao, Y., Fu, L., and Li, W. (2010). CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26, 680–682. doi: 10.1093/BIOINFORMATICS/BTQ003
Jamieson, A. J. (2015). The Hadal Zone: Life in the Deepest Oceans. Cambridge: Cambridge University Press.
Jamieson, A. J., Fujii, T., Mayor, D. J., Solan, M., and Priede, I. G. (2010). Hadal trenches: the ecology of the deepest places on Earth. Trends Ecol. Evol. 25, 190–197. doi: 10.1016/j.tree.2009.09.009
Jamieson, A. J., Fujii, T., Solan, M., and Priede, I. G. (2009). HADEEP: free-falling landers to the deepest places on Earth. Mar. Technol. Soc. J. 43, 151–160. doi: 10.4031/MTSJ.43.5.17
Jamieson, A. J., Kilgallen, N. M., Rowden, A. A., Fujii, T., Horton, T., Lörz, A. N., et al. (2011). Bait-attending fauna of the Kermadec Trench, SW Pacific Ocean: evidence for an ecotone across the abyssal-hadal transition zone. Deep Sea Res. Part I Oceanogr. Res. Pap. 58, 49–62. doi: 10.1016/j.dsr.2010.11.003
Jamieson, A. J., Lacey, N. C., Lörz, A. N., Rowden, A. A., and Piertney, S. B. (2013). The supergiant amphipod Alicella gigantea (Crustacea: Alicellidae) from hadal depths in the Kermadec Trench, SW Pacific Ocean. Deep Sea Res. Part II Top. Stud. Oceanogr. 92, 107–113. doi: 10.1016/j.dsr2.2012.12.002
Jamieson, A. J., and Linley, T. D. (2021). Hydrozoans, scyphozoans, larvaceans and ctenophores observed in situ at hadal depths. J. Plankton Res. 43, 20–32. doi: 10.1093/plankt/fbaa062
Jamieson, A. J., Stewart, H. A., Weston, J. N. J., and Bongiovanni, C. (2021b). Hadal fauna of the South Sandwich Trench, Southern Ocean: baited camera survey from the Five Deeps Expedition. Deep Sea Res. Part II Top. Stud. Oceanogr. 194:104987. doi: 10.1016/J.DSR2.2021.104987
Jamieson, A. J., Linley, T. D., Eigler, S., and Macdonald, T. (2021a). A global assessment of fishes at lower abyssal and upper hadal depths (5000 to 8000m). Deep Sea Res. Part I Oceanogr. Res. Pap. 178:103642. doi: 10.1016/J.DSR.2021.103642
Jazdzewska, A. M., Horton, T., Hendrycks, E. A., Mamos, T., Driskel, A., Brix, S., et al. (2021). Pandora’s box in the deep sea-intraspecific diversity patterns and distribution of two congeneric scavenging amphipods. Front. Mar. Sci. 8:750180. doi: 10.3389/fmars.2021.750180
Jażdżewska, A. M., and Mamos, T. (2019). High species richness of Northwest Pacific deep-sea amphipods revealed through DNA barcoding. Prog. Oceanogr. 178:102184. doi: 10.1016/j.pocean.2019.102184
Kamenskaya, O. E. (1977). Two new species of ultraabyssal amphipods from Yap Trench. Akad. Nauk S.S.S.R. 108, 105–114.
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/BIB/BBX108
Kilgallen, N. M. (2015). Three new species of Hirondellea (Crustacea, Amphipoda, Hirondelleidae) from hadal depths of the Peru-Chile Trench. Mar. Biol. Res. 11, 34–48. doi: 10.1080/17451000.2014.889309
Kilgallen, N. M., and Lowry, J. K. (2015). A review of the scopelocheirid amphipods (Crustacea, Amphipoda, Lysianassoidea), with the description of new taxa from Australian waters. Zoosyst. Evol. 91, 1–43. doi: 10.3897/zse.91.8440
Kniesz, K., Jażdżewska, A. M., Martínez Arbizu, P., and Kihara, T. C. (2022). DNA barcoding of scavenging amphipod communities at active and inactive hydrothermal vents in the Indian Ocean. Front. Mar. Sci. 8:752360. doi: 10.3389/FMARS.2021.752360
Kobayashi, H., Hatada, Y., Tsubouchi, T., Nagahama, T., and Takami, H. (2012). The hadal amphipod Hirondellea gigas possessing a unique cellulase for digesting wooden debris buried in the deepest seafloor. PLoS One 7:e42727. doi: 10.1371/journal.pone.0042727
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/MOLBEV/MSY096
Lacey, N. C., Mayor, D. J., Linley, T. D., and Jamieson, A. J. (2018). Population structure of the hadal amphipod Bathycallisoma (Scopelocheirus) schellenbergi in the Kermadec Trench and New Hebrides Trench, SW Pacific. Deep Sea Res. Part II Top. Stud. Oceanogr. 155, 50–60. doi: 10.1016/j.dsr2.2017.05.001
Lacey, N. C., Rowden, A. A., Clark, M. R., Kilgallen, N. M., Linley, T., Mayor, D. J., et al. (2016). Community structure and diversity of scavenging amphipods from bathyal to hadal depths in three South Pacific Trenches. Deep Sea Res. Part I Oceanogr. Res. Pap. 111, 121–137. doi: 10.1016/j.dsr.2016.02.014
Leduc, D., and Wilson, J. (2016). Benthimermithid nematode parasites of the amphipod Hirondellea dubia in the Kermadec Trench. Parasitol. Res. 115, 1675–1682. doi: 10.1007/s00436-016-4907-7
Li, W., and Godzik, A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. doi: 10.1093/BIOINFORMATICS/BTL158
Lowry, J. K., and Stoddart, H. E. (2010). The deep-sea scavenging genus Hirondellea (Crustacea: Amphipoda: Lysianassoidea: Hirondelleidae fam. nov.) in Australian waters. Zootaxa 2329, 37–55. doi: 10.11646/ZOOTAXA.2329.1.3
Monaghan, M. T., Wild, R., Elliot, M., Fujisawa, T., Balke, M., Inward, D. J. G., et al. (2009). Accelerated species inventory on Madagascar using coalescent-based models of species delineation. Syst. Biol. 58, 298–311. doi: 10.1093/SYSBIO/SYP027
Page, T. J., Choy, S. C., and Hughes, J. M. (2005). The taxonomic feedback loop: symbiosis of morphology and molecules. Biol. Lett. 1, 139–142. doi: 10.1098/RSBL.2005.0298
Palumbi, S. R., Martin, A., Romano, S., McMillan, W. O., Stice, L., and Grabowski, G. (1991). The Simple Fool’s Guide to PCR, Version 2.0. Honolulu, HI: University of Hawaii.
Pons, J., Barraclough, T. G., Gomez-Zurita, J., Cardoso, A., Duran, D. P., Hazell, S., et al. (2006). Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 55, 595–609. doi: 10.1080/10635150600852011
Puillandre, N., Lambert, A., Brouillet, S., and Achaz, G. (2012). ABGD, automatic barcode gap discovery for primary species delimitation. Mol. Ecol. 21, 1864–1877. doi: 10.1111/J.1365-294X.2011.05239.X
R Core Team (2020). R: A Language and Environment for Statistical Computing. Available online at: https://www.R-project.org/ (accessed October 25, 2021).
Ratnasingham, S., and Hebert, P. D. N. (2007). BOLD: the barcode of life data system (http://www.barcodinglife.org). Mol. Ecol. Notes 7, 355–364.
Ratnasingham, S., and Hebert, P. D. N. (2013). A DNA-based registry for all animal species: the Barcode Index Number (BIN) system. PLoS One 8:e66213. doi: 10.1371/JOURNAL.PONE.0066213
Ritchie, H., Jamieson, A. J., and Piertney, S. B. (2015). Phylogenetic relationships among hadal amphipods of the Superfamily Lysianassoidea: implications for taxonomy and biogeography. Deep Sea Res. Part I Oceanogr. Res. Pap. 105, 119–131. doi: 10.1016/j.dsr.2015.08.014
Stewart, H. A., and Jamieson, A. J. (2018). Habitat heterogeneity of hadal trenches: considerations and implications for future studies. Prog. Oceanogr. 161, 47–65. doi: 10.1016/j.pocean.2018.01.007
Suchard, M. A., Lemey, P., Baele, G., Ayres, D. L., Drummond, A. J., and Rambaut, A. (2018). Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4, doi: 10.1093/VE/VEY016
Taylor, M. L., and Roterman, C. N. (2017). Invertebrate population genetics across Earth’s largest habitat: the deep-sea floor. Mol. Ecol. 26, 4872–4896. doi: 10.1111/mec.14237
The Inkscape Team (2017). Inkscape 0.92.2. Available online at: https://inkscape.org/release/0.92.2/windows/ (accessed October 25, 2021)
Weston, J. N. J., Peart, R. A., Stewart, H. A., Ritchie, H., Piertney, S. B., Linley, T. D., et al. (2021b). Scavenging amphipods from the Wallaby-Zenith Fracture Zone: extending the hadal paradigm beyond subduction trenches. Mar. Biol. 168:1. doi: 10.1007/s00227-020-03798-4
Weston, J. N. J., Espinosa-Leal, L., Wainwright, J. A., Stewart, E. C. D., González, C. E., Linley, T. D., et al. (2021a). Eurythenes atacamensis sp. nov. (Crustacea: Amphipoda) exhibits ontogenetic vertical stratification across abyssal and hadal depths in the Atacama Trench, eastern South Pacific Ocean. Mar. Biodivers. 51:51. doi: 10.1007/s12526-021-01182-z
Weston, J. N. J., Peart, R. A., and Jamieson, A. J. (2020). Amphipods from the Wallaby-Zenith Fracture Zone, Indian Ocean: new genus and two new species identified by integrative taxonomy. Syst. Biodivers. 18, 57–78. doi: 10.1080/14772000.2020.1729891
Wilson, J. P. A., Schnabel, K. E., Rowden, A. A., Peart, R. A., Kitazato, H., and Ryan, K. G. (2018). Bait-attending amphipods of the Tonga Trench and depth-stratified population structure in the scavenging amphipod Hirondellea dubia Dahl, 1959. PeerJ 7:e5994. doi: 10.7717/peerj.5994
Wolff, T. (1959). The hadal community, an introduction. Deep Sea Res. 6, 95–124. doi: 10.1016/0146-6313(59)90063-2
Keywords: Lysianassoidea, integrative taxonomy, bathymetric distributions, biogeography, deep sea, scavenging amphipods, subduction trenches
Citation: Weston JNJ and Jamieson AJ (2022) The Multi-Ocean Distribution of the Hadal Amphipod, Hirondellea dubia Dahl, 1959 (Crustacea, Amphipoda). Front. Mar. Sci. 9:824640. doi: 10.3389/fmars.2022.824640
Received: 29 November 2021; Accepted: 27 January 2022;
Published: 04 March 2022.
Edited by:
Eleonora Puccinelli, Université de Bretagne Occidentale, FranceCopyright © 2022 Weston and Jamieson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Johanna N. J. Weston, johanna.weston@ncl.ac.uk
†ORCID: Johanna N. J. Weston, orcid.org/0000-0002-7142-3081; Alan J. Jamieson, orcid.org/0000-0001-9835-2909
 Johanna N. J. Weston
Johanna N. J. Weston Alan J. Jamieson
Alan J. Jamieson