Stirring the Deep, Disentangling the Complexity: Report on the Third Species of Thermochiton (Mollusca: Polyplacophora) From Haima Cold Seeps
- 1School of Marine Science and Engineering, Qingdao Agricultural University, Qingdao, China
- 2Laboratory of Marine Organism Taxonomy and Phylogeny, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 3Key Laboratory of Ocean Circulation and Waves, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 4Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao, China
- 5Laboratory for Ocean Dynamics and Climate, Pilot National Laboratory for Marine Science and Technology, Qingdao, China
- 6Zoological Institute, Russian Academy of Sciences, St. Petersburg, Russia
This study documents a new deep-sea chiton from the Haima cold seeps. Thermochiton xui. nov. is the third species of the genus Thermochiton and the first occurrence of this genus in the South China Sea. This species is identified by its morphological characteristics and the molecular sequence of a Thermochiton species is reported for the first time. The placement of the new species is determined in the phylogenetic tree of Ischnochitonidae by Maximum Likelihood (ML) and Bayesian inference (BI) methods, based on the sequences of the mitochondrial cytochrome c oxidase subunit I (COI), 16S ribosomal DNA (16S), and nuclear 28S ribosomal DNA (28S) gene regions. Bayesian evolutionary analysis with an uncorrelated relaxed clock approach indicated that this new species is estimated to have diverged from its most closely related shallow-water ischnochitonid taxa 5.10–10.07 million years ago in the Late Miocene. A regional ocean general circulation model was used to estimate the potential dispersal ability of the three species of Thermochiton. Because it is highly unlikely for one species to have spread between the northwest and southwest Pacific to the localities in which this genus has been found to date, we propose that ‘stepping-stone’ habitats and/or ‘bridge species’ were involved in the dispersal and evolution of these cold-seep endemic chitons.
The ZooBank Life Science Identifier (LSID) for this publication is: urn:lsid:zoobank.org:pub:AD93E4BC-2977-405E-B681-D956C5C66D83. And the ISID for Thermochiton xui sp. nov. is: urn:lsid:zoobank.org:act:0C75D2E3-F30E-4970-9BC2-3363B397720C.
Introduction
The Haima cold-seep field, covering 618 km2, was recently discovered on the northwestern slope of the South China Sea. Within this field, an area of 350 km2 has had cold seep activity and developed into cold-seep carbonate rocks with crusts and nodules that harbor a distinct biological community (Liang et al., 2017; Zhao et al., 2020). To date, more than 80 species of macrobenthic organisms have been sampled from the Haima cold seeps, including mostly mollusks, polychaetes, and crustaceans, a few species of echinoderms, and other animals. Since the discovery of the Haima cold seeps, many new molluskan species and the first recorded occurrences of species in the South China Sea have been reported while exploring the benthic communities associated with the cold seeps (Zhang et al., 2016; Zhang and Zhang, 2017; Chen et al., 2018; Sun et al., 2018; Jiang et al., 2019; Xu et al., 2019; Dong et al., 2021). Among them, primarily gastropods and bivalves have been found, with only one reported species of the class Polyplacophora (Dong et al., 2021).
Most polyplacophorans, also called chitons, live in shallow waters, usually in the intertidal zones on rocky substrates. Only a small fraction of chitons have been found in abyssal or bathyal habitats, especially in chemosynthetic environments such as sunken wood, cold seeps, or hydrothermal vents. Until 2010, only 18 chiton species had been found in chemosynthetic areas, of which 16 species were found in cold seeps (Schwabe and Sellanes, 2010). However, new deep-sea species have been discovered with the improvement of exploration technology (Sigwart et al., 2010; Sigwart and Sirenko, 2012; Sirenko and Sellanes, 2016; Sigwart and Chen, 2018; Sirenko, 2020). Saito and Okutani (1990) described a chiton of the order Chitonida from hydrothermal vents in the central Okinawa Trough, East China Sea. They established the genus Thermochiton for Thermochiton undocostatus Saito & Okutani, 1990 and placed the genus in the family Ischnochitondae. The second member of the genus, Thermochiton papuaensis, was reported in deep waters near Papua New Guinea, likely inhabiting communities with chemical activity (Sirenko, 2020). However, both species were described merely by their morphology without any genetic information. Their relationship within the family Ischnochitondae has not been determined by molecular analysis.
Ischnochitonidae is the largest family of Polyplacophora. Kaas and Van Belle (1980) classified the family into 18 genera, comprising approximately 400 species, which contained almost half of the known chitons at the time. Although the taxa were later rearranged (Sirenko, 2006), this large family currently includes 10 genera and approximately 228 species (World Register of Marine Species). Chitons of this family have distinct apophyses, well-developed slits on all valves, insertion plates that are not pectinated, and dorsal surfaces that lack ocelli. Their adanal gills do not extend to the anus. Additionally, all members of the family Ischnochitonidae have a scaly dorsal girdle and varying tegmental sculptures (Kaas and Van Belle, 1987). The inherent paraphyly and lack of well-defined diagnostic morphological features of this family have led to taxonomic controversy. Some molecular studies suggest that the family Ischnochitonidae is not monophyletic (Okusu et al., 2003; Owada, 2018; Irisarri et al., 2020). Even the monophyly of its type genus, Ischnochiton, remains doubtful (Owada, 2018; Irisarri et al., 2020). However, most ischnochitonids have only been described by their morphology, and molecular information is scarce. Indeed, the sequences of mitochondrial cytochrome c oxidase subunit I (COI), the most widely used effective DNA barcoding gene marker, have only been published for 24 species of Ischnochitonidae in the NCBI database.
The evolutionary history of geographically separated but closely related species is tangled but fascinating. Chemoautotrophic environments are ideal natural laboratories for studying the spread, connectivity, and evolution. Hydrothermal vents or cold seeps, are essentially isolated islands in the deep sea. As most vent-endemic species are benthic with limited mobility at their adult stage, the larval stage is vital for dispersal and colonization of species obligately living in chemoautotrophic environments (Tyler and Young, 1999; Tyler and Young, 2003). The wide geographic range of distributed species and long-distance dispersal or high connectivity of panmictic populations are achieved via long periods of planktonic larval duration (PLD) for swimming larvae (Adams et al., 2012; Fukumori and Kano, 2014; Yahagi et al., 2019). Some vent or seep mollusks, such as Shinkailepas myojinensis (Yahagi et al., 2017), Shinkailepas briandi (Yahagi et al., 2019), Bathymodiolus childressi, and Thalassonerita naticoidea (= Bathynerita naticoidea) (Arellano et al., 2014), develop long-lived planktotrophic larvae (Warén and Bouchet, 2001; Fukumori and Kano, 2014), which can vertically migrate hundreds of meters to photic and even euphotic zones to hunt for abundant food supplies (Bouchet and Warén, 1994; Arellano et al., 2014). These larvae were considered to float for at least 7–12 months in the pelagic water column before settling, based on the sizes of planktonic and settled larvae, as well as post-settlement juveniles (Arellano et al., 2014; Yahagi et al., 2017). The longer PLD of these species and stronger currents at the sea surface presumably fuel broad-ranged distribution and genetic homogeneity (Bouchet and Warén, 1994; Yahagi et al., 2017). Several crustaceans have developed the same strategy (Teixeira et al., 2011; Beedessee et al., 2013). However, the strategies employed by vent-associated chitons to colonize new territories and expand their distribution have not yet been reported, partly because of their rare occurrence.
During an exploratory cruise of the Haima cold seeps, two specimens of deep-sea chiton were collected and reported as Leptochiton tenuidontus Saito & Okutani, 1990 by Dong et al. (2021). This species was misidentified and should be classified as a new species of the genus Thermochiton. In this study, we clarify the identification of this new species. Our account is based on analyzing the valve, girdle, and radula morphological characteristics of the chiton, as well as DNA sequencing of the mitochondrial COI, 16S ribosomal DNA (16S), and nuclear 28S ribosomal DNA (28S) gene regions. DNA sequences were used to confirm the arrangement of this Thermochiton species within the family Ischnochitondae and to explore its evolutionary relationship with its shallow-water relatives. Further, a numerical model was used to simulate the dispersal range and estimate the potential dispersal ability, thus suggesting the evolutionary pathway of Thermochiton species.
Material and Methods
Sample Collection
The samples for this study were obtained during a cruise to the Haima cold seeps in the South China Sea on 19 May 2018 (Figure 1). The two individual specimens were directly preserved in 75% ethanol. The specimens were initially identified at the species level based on their morphology according to identification guidelines (Saito and Okutani, 1990; Saito, 2004; Saito et al., 2008; Sirenko, 2021). The specimens were deposited in the Marine Biological Museum, Chinese Academy of Sciences (MBMCAS), Qingdao, China.
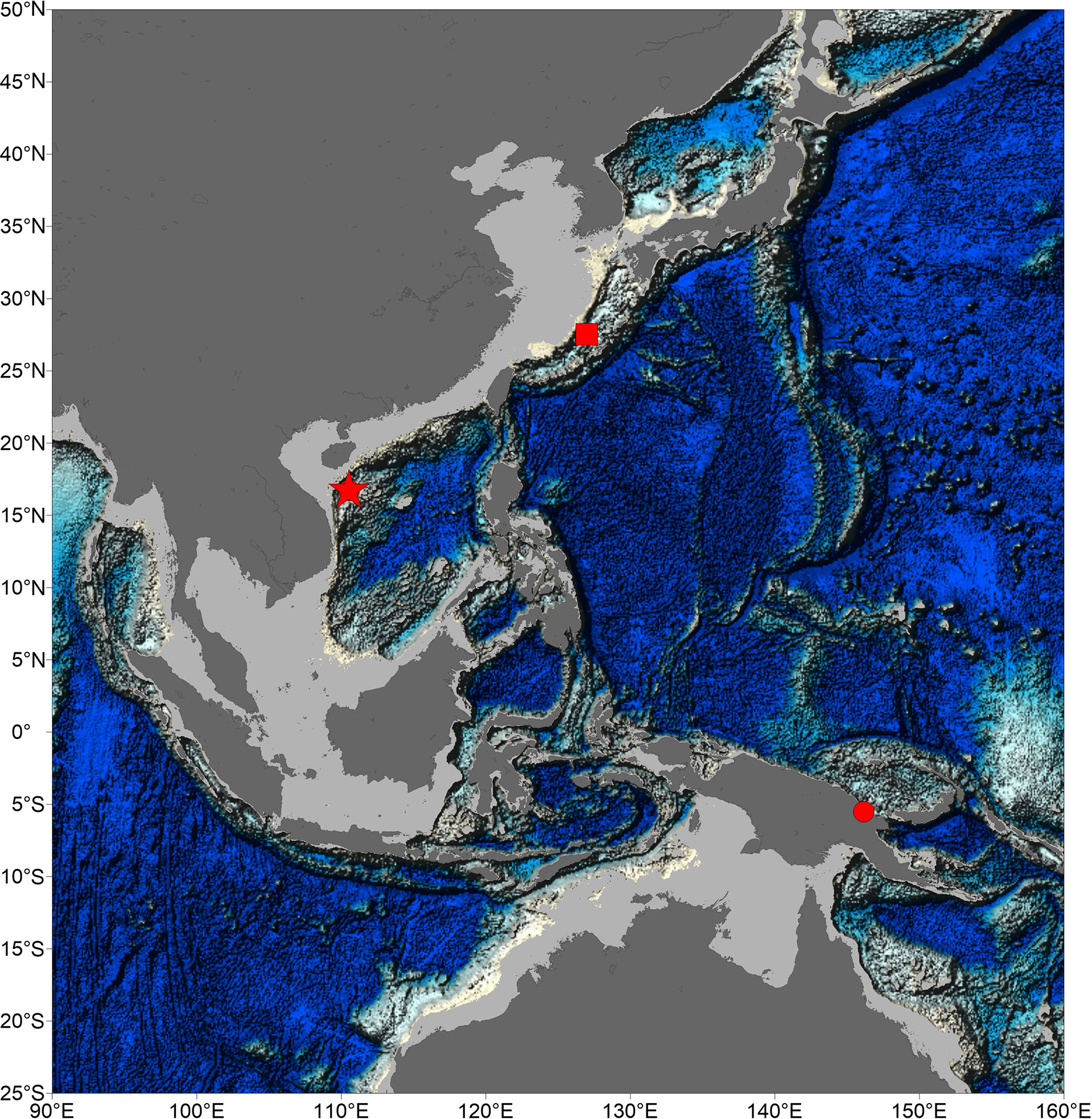
Figure 1 Map of samples sites of species of genus Thermochiton. Location of Haima cold seeps was indicated by a pentacle. Type localities of Thermochiton undocostatus and Thermochiton papuaensis were indicated by square and circle, respectively.
Morphological Observations
The two specimens were identified at the species level under a Zeiss SteREO Discovery.V12 stereo microscope (Zeiss, Wetzlar, Germany). The body length and width were measured, and the number of gills and gill types were observed. Specimens were placed in 7% NaOH and boiled for 10 min, followed by boiling twice in freshwater. Selected valves (head plate, 5th or 2nd plate of the intermediate plates, and tail plate), half of the radula, and a portion of the girdle were glued to the sample stage with conductive glue, gold plated in a vacuum sputter coater, and examined under a Hitachi S-3400N scanning electron microscope (Hitachi, Tokyo, Japan) at an accelerating voltage of 5 kV.
Genetic Study
Total DNA was extracted using a marine animal genomic DNA extraction kit (Tiangen Biotechnology, Beijing, China) according to the instructions of the manufacturer. To explore the genetic distance and relationship of the species to common coastal and deep-sea chitons of the family Ischnochitonidae, the mitochondrial COI, 16S rRNA (16S), and nuclear 28S rRNA (28S) gene regions were sequenced and analyzed.
Polymerase chain reaction (PCR) amplification of the mitochondrial COI gene was carried out using primers LCO1490 (5′-GGTCAACAAATCATAAAGATATTGG-3′) and HCO2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′). The mitochondrial 16S rRNA gene was amplified using primers 16Sa (5’-CGCCTGTTTATCAAAAACAT-3’) and 16Sb (5’-CTCCGGTTTGAACTCAGATCA-3’). The nuclear 28S rRNA gene was amplified using primers 28Sa (5’-GACCCGTCTTGAAACACGGA-3’) and 28Sb (5’-TCGGAAGGAACCAGCTAC-3’). PCR amplifications were carried out using a total reaction volume of 25 μl, with 12 μl 2×Es Taq MasterMix (CoWin Biosciences Co., Ltd., Beijing, China), 2 µl of template DNA (50 ng/µl), 1 µl of each primer (10 M), and 9 µl of dH2O. The thermal cycling conditions for PCR amplification were as follows: initial denaturation at 95°C for 3 min, followed by 35 cycles of 95°C (30 s), annealing temperature of 50°C for 1 min, extension at 72°C for 1 min, and a final elongation at 72°C for 10 min. Double-stranded PCR products were purified and sequenced by BGI Tech Solutions Co., Ltd. PCR products were detected by an agarose gel.
The 24 species of the family Ischnochitonidae with available COI, 16S rRNA, and 28S rRNA gene sequences were also included in the analysis (Table 1). Callistochiton jacobaeus and Callistochiton antiquus of the family Callistoplacidae, Rhyssoplax kurodai of the family Chitonidae, and Chaetopleura apiculata and Chaetopleura angulata of the family Chaetopluridae were selected as outgroups based on their current taxonomic relationships.
All sequences were aligned using MAFFT v.7 software (Katoh and Standley, 2013) with the G-INS-I algorithm. Alignments were carried out by eye and BLAST analyses of the NCBI database. The online version of Automatic Barcode Gap Discovery (ABGD) (Puillandre et al., 2012) was used to detect barcode gaps and help support species delineation (https://bioinfo.mnhn.fr/abi/public/abgd/abgdweb.html) using four randomly selected COI sequences of the 24 available species of Ischnochitonidae obtained from published studies, excluding outgroups (Supplementary Table S1). The analysis settings were as follows: Pmin = 0.001, Pmax = 0.1, Steps = 10, X (relative gap width) = 1.0. Then, the Kimura two-parameter model was used to measure a matrix of pairwise distances (Kimura, 1980). The COI, 16S, and 28S gene sequences from the same individual were concatenated using SequenceMatrix software (Vaidya et al., 2011) to conduct phylogenetic analysis of a combined-gene data set. Best-fitting sequence evolution models were calculated using jModelTest 2.1.10 software (Darriba et al., 2012), deriving the best-fit general time-reversible (GTR)+G model. The maximum likelihood (ML) method was executed using RaxML1.5b1 software (Stamatakis, 2014). The test was executed 1,000 times with the default settings. The support bootstrap scores of nodes were obtained from the best scoring tree after 1,000 bootstrap replications. A Bayesian phylogenetic analysis was carried out using MrBayes v.3.2.6 software (Ronquist et al., 2012). The program was run for 10,000,000 generations, sampling every 1,000 generations.
The uncorrelated relaxed clock method was used to estimate the divergence times of the different taxa using BEAST v. 1.10.4 software, based on the Bayesian interference (BI) tree of the concatenated sequences of three genes (Suchard et al., 2018). The earliest fossil record of Mopalia, 15–17.2 million years ago (MYA) (Kelly and Eernisse, 2008), was taken as the calibration point. An uncorrelated relaxed lognormal clock was derived for each partition using the GTR substitution model, assuming that the tree prior had a Yule species formation process. This analysis was run for 100 million generations, sampling every 1,000 generations with a 10% burn-in rate. The final result image was rendered using Figtree v1.4.4 software.
Tracer Simulation
The diffusion and advection processes of the biological tracer in the ocean were simulated using a regional ocean general circulation model (OGCM), which was constructed based on the Massachusetts Institute of Technology general circulation model (MITgcm) (Marshall et al., 1997). Our model domain ranged from 25°S to 45°N and from 110°E to 174°E, with a horizontal resolution of 1/10° × 1/10° and 82 z-levels in the vertical. Vertical mixing was represented using the K-Profile Paremeterization (KPP) scheme (Large et al., 1994), and background vertical viscosity and diffusivity were set as 10−4 and 10−5 m2 s−1, respectively. The potentially unresolved eddy processes were represented by the parameterizations described by Gent and McWilliams (1990) and Redi (1982), with both layer thickness and isopycnal diffusivity set at 500 m2 s−1. The initial temperature and salinity of the field were obtained from the climatological monthly mean data for January, which were derived from the Generalized Digital Environmental Model, version 3 (GDEMv3; http://www.usgodae.org/pub/outgoing/static/ocn/gdem/) dataset. The climatological monthly mean wind stress was calculated from the cross-calibrated multiplatform (CCMP) datasets (Atlas et al., 2011), with a relaxation of the sea surface temperature and salinity to the monthly mean GDEMv3 climatology.
The model was started from an initial state of rest and ran for 100 years. The simulated total kinetic energy of the upper ocean layer reached a quasi-equilibrium state after a 100-year run. Subsequently, the simulated monthly mean temperature, salinity, and currents of the last month were used as the initial states of the following tracer experiments. The horizontal and vertical diffusivities were the same as those in the circulation model. The three localities of Thermochiton species were selected as the origins of the model: (1) the central Okinawa Trough, the type locality of T. undocostatus Saito et Okutani, 1990 (Saito and Okutani, 1990); (2) Papua New Guinea, the type locality of T. papuaensis Sirenko, 2020 (Sirenko, 2020); and (3) the Haima cold seeps in the South China Sea, where our specimens were collected. A tracer experiment was carried out for each origin. At the corresponding model grid of each origin, 100 million floats/m3 were released at the first model level of 2.5 m water depth. All three experiments ran for three years, with the monthly-mean tracer concentration fields used for analysis.
Results
Systematics
Class Polyplacophora Gray, 1821
Order Chitonida Thiele, 1909
Family Ischnochitonidae Dall, 1889
Genus Thermochiton Saito et Okutani, 1990
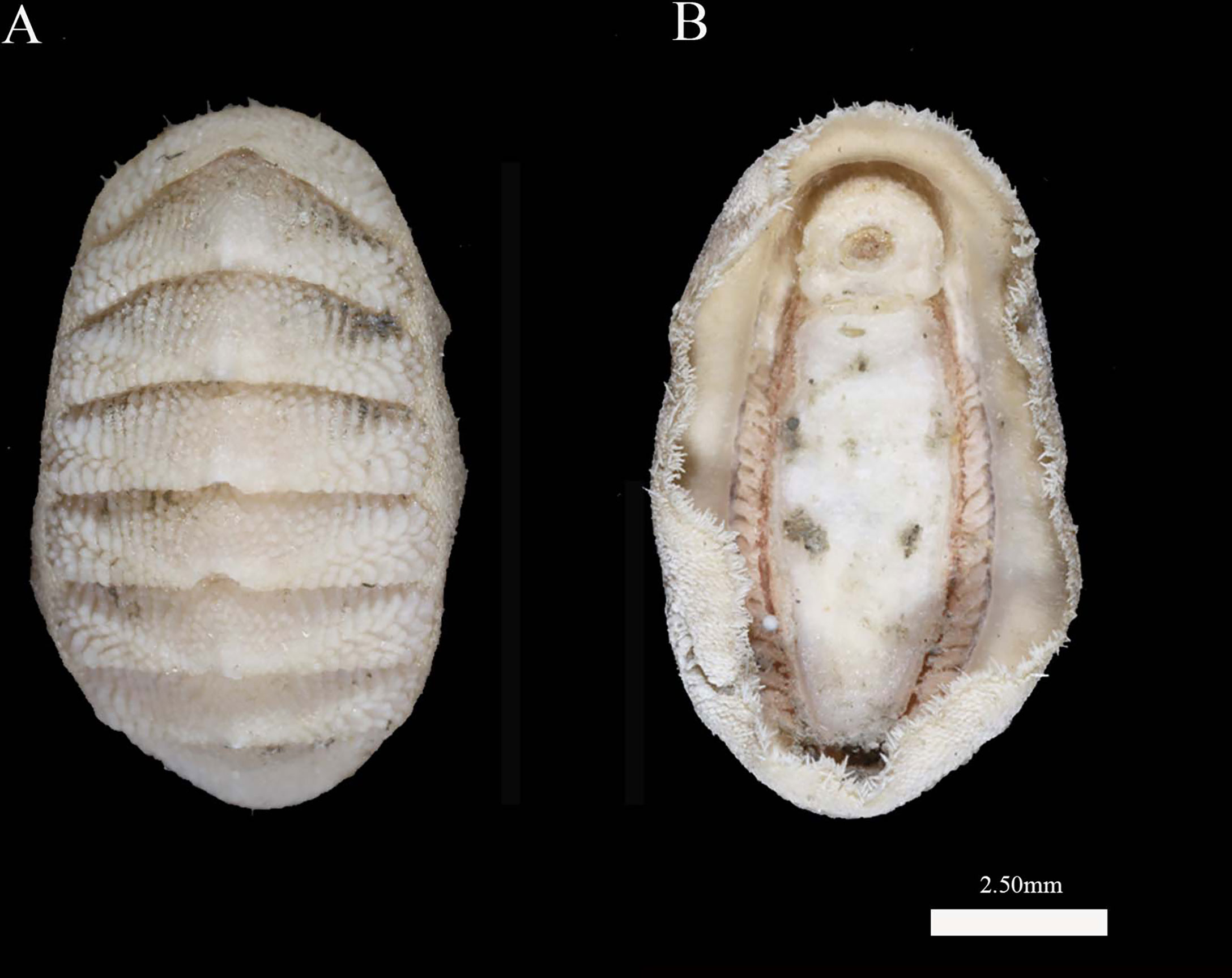
Thermochiton xui Wang, Zhang & Sirenko sp. nov. (Figures 2–5)
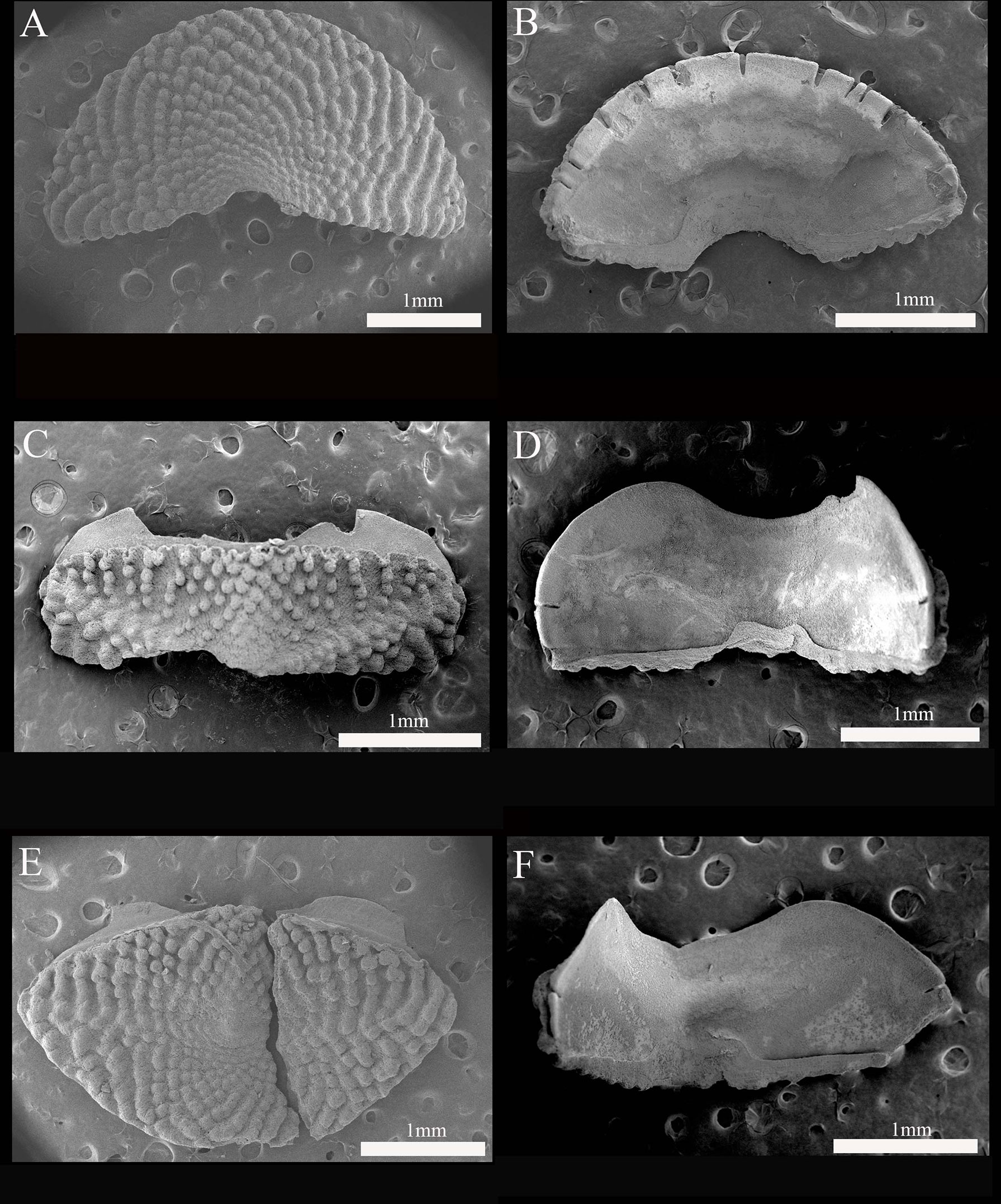
Figure 3 Thermochiton xui sp. nov. holotype: MBM229030. (A) Head valve, dorsal view; (B) Head valve, ventral view; (C) Valve III, dorsal view; (D) Valve III, ventral view; (E) Tail valve, dorsal view; and (F) Valve IV, ventral view.
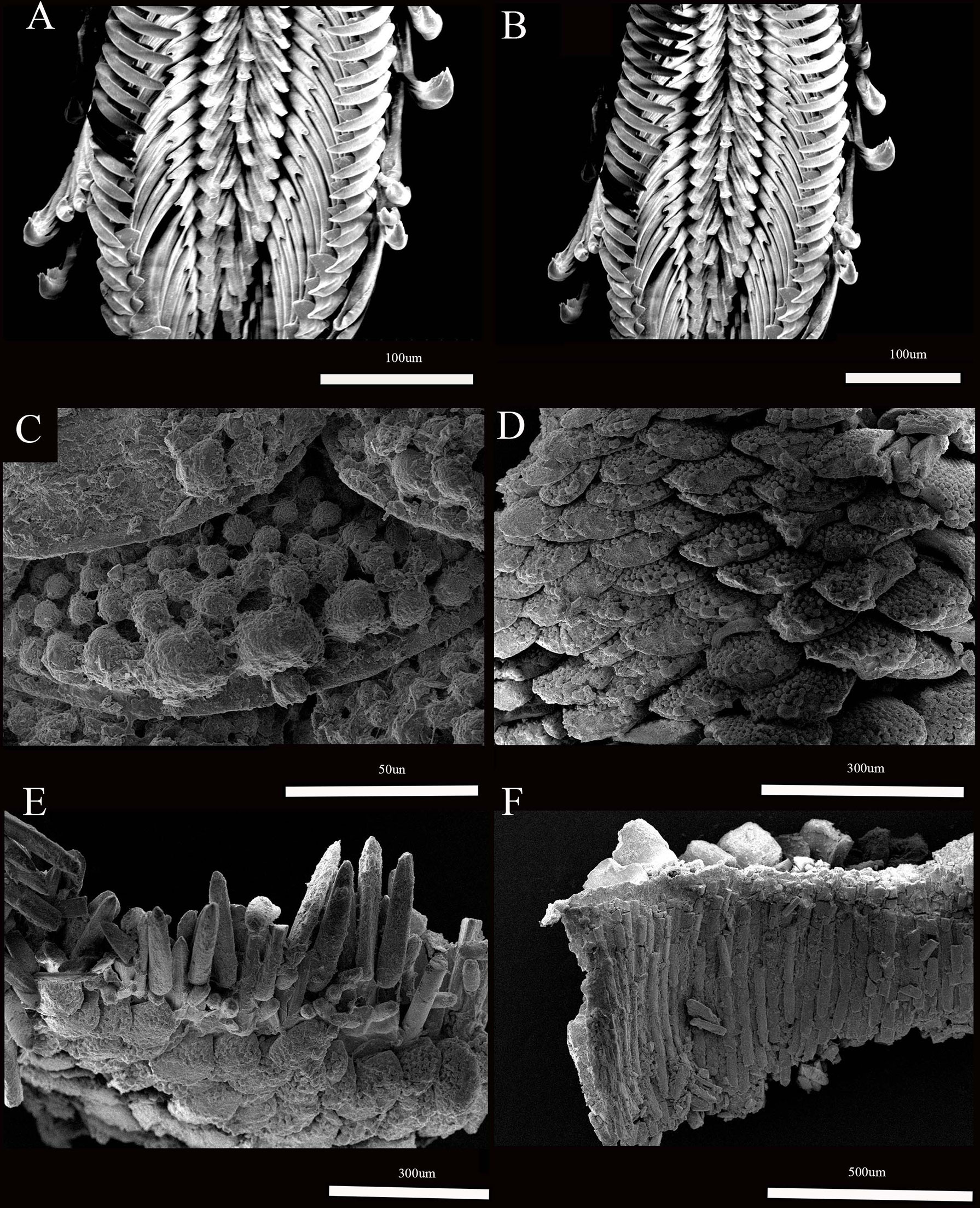
Figure 4 Thermochiton xui sp. nov. paratype: MBM229031. (A, B) Portion of radula; (C, D) Dorsal scales; (E) Dorsal scales and marginal spicules; and (F) ventral scales.
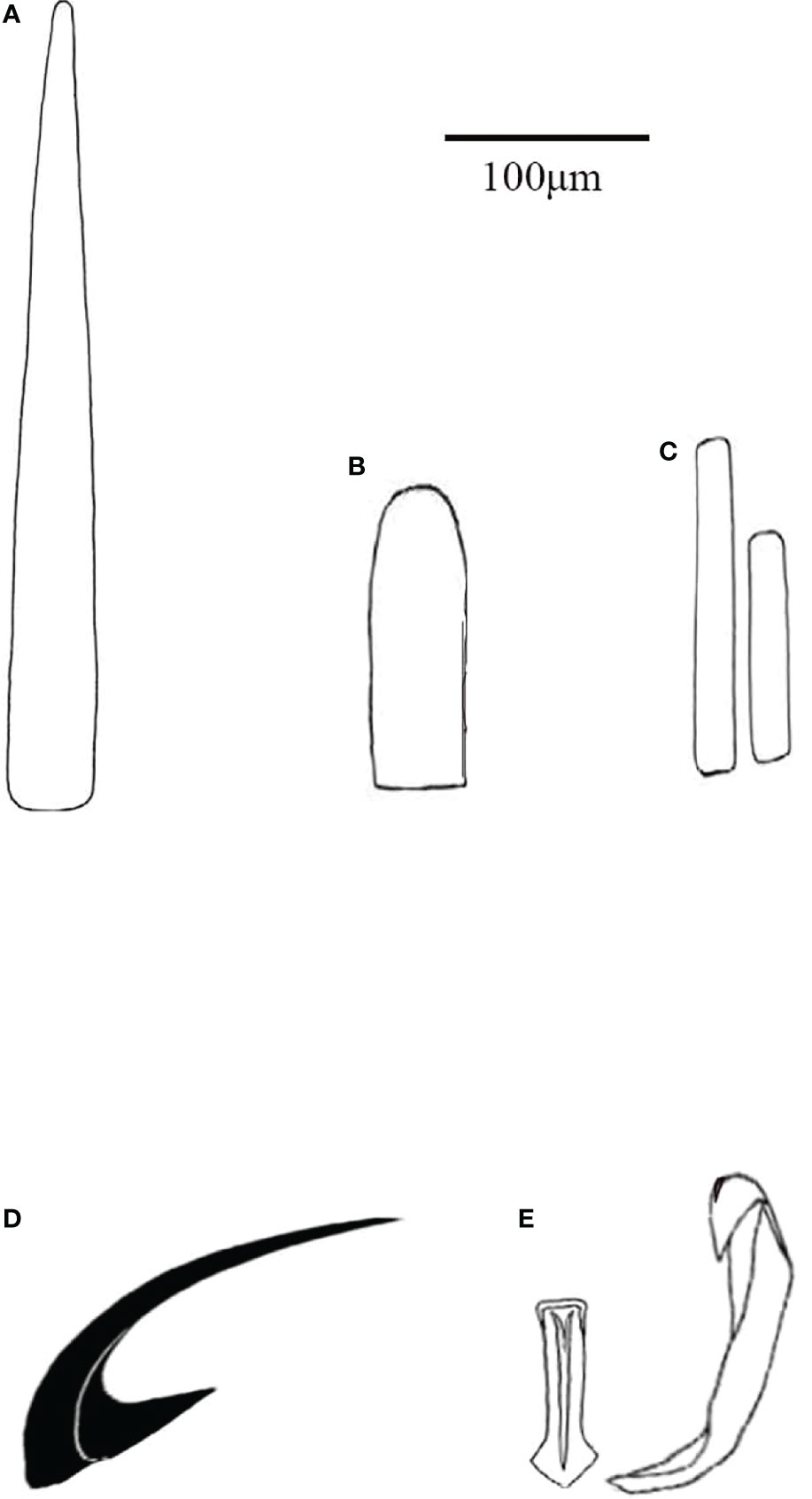
Figure 5 Thermochiton xui sp. nov. (A) Smooth marginal needle; (B) Short blunt-topped marginal spicules; (C) Ventral scales; (D) Head of major lateral tooth of radula. and (E) Central and first lateral teeth of radula.
Type Material
Holotype: MBM229030, BL 9.0 mm. Paratype: MBM229031, BL 8.5 mm. All collected from type locality, May 19, 2018, and deposited in the Marine Biological Museum, Chinese Academy of Sciences (MBMCAS), Qingdao.
Type Locality
Haima cold seeps, South China Sea, off southern Hainan Island.
Distribution and Habitat
Only known from the type locality, Haima cold seeps, South China Sea, chemosynthetic habitat at a depth of 1,380–1,390 m.
Etymology
This species is named after Prof. Fengshan Xu for his contributions to Chinese malacology on Polyplacophora. He is the first Chinese author who has published the taxonomy of chitons and has described one genus, Sinolorica (= Loricella Pilsbry, 1893) and two species of chiton, Sinolorica scissurata (= Loricella scissurata) and Deshayesiella sinica (= Hanleya sinica) from the East China Sea (Xu, 1990).
Diagnosis
Animal small, oval to elongated oval in shape, dorsal elevation rather high, and subcarinated. The valves rather elevated, not beaked. Tegmentum white in color. Slit formula 11/1(2)/12. Girdle moderately wide and dorsally covered with dense, imbricating, obliquely set, squarish, curved, and wide scales with large round granules at the distal end and sculpted into approximately 18 longitudinal rows of smaller granules on the dorsal side of the scales. Two types of marginal spicules long, smooth, needle-like spicules and short, smooth, blunt-topped spicules. Central tooth of the radula elongated with a wide base. Centro-lateral tooth rough, wedge-shaped, attenuated posteriorly, taller than the central tooth, with a subtriangular blade at the antero-dorsal edge. The top of the lateral central tooth narrow with a sharp, oblique, inward tip. The head of the major lateral tooth of the radula is unicuspidal, bent, sickle-shaped, with an extended base ending with a long, sharp-pointed process directed inside.
Description
Animal elliptical in shape, with a body length of 9.0 mm. Valves rather elevated valves, not beaked, and the side slopes weakly convex. Color of the valves and girdle is white with friable, rusty brown deposits.
Head valve is semicircular, with a straight front slope, and the posterior margin is widely V-shaped and weakly notched in the middle. Intermediate valves rather short compared to their width, rectangular, with the front margin slightly convex, side margins rounded, the front margin slightly angular, the hind margin straight, with no beaks, and lateral areas strongly raised. Tail valve less than semicircular, front margin more or less angular, less wide than the head valve, the length is about half the width, the antemucronal and postmucronal slopes slightly convex, and the mucro central and not swollen.
All the valves in the jugal area delicately sculptured by randomly arranged, oval, flattened granules. Pleural area of intermediate valves covered with randomly arranged, large, and drop-shaped granules. Lateral area covered with elongated granules formed undulating costae. Numerous pores of aesthetes evenly covering both the costae and the interstices between them.
Articulamentum very thin, transparent, and glassy white; apophyses short, very wide, and rounded, coalescing across the small, shallow sinus; slit formula of insertion plates 11/1(2)/12; slit rays barely visible; and the solid teeth short, blunt, and the eaves are narrow. Four of the six intermediate valves with 2 slits on one side, whereas the other sides and other valves with only one slit per side. Ventral tegmental callus well developed.
Girdle rather wide, colored like the tegmentum, and dorsally covered with imbricating obliquely set, curved, wide scales, with large round granules at the distal end and about 18 longitudinal rows of smaller granules on the dorsal side of the scales. Scales near valves straight, but in other places angled in different directions. Some scales with 1–4 short longitudinal ribs near the side margin. Marginal spicules of two types: long, smooth spicules-needles and short, smooth blunt-topped spicules. Ventral side of the girdle is paved with smooth, elongate scales.
Radula 4.1 mm long with 110 transverse rows of mature teeth. Central tooth rectangular, oblong, parallel-sided, keeled medially, with a small blade and a large base. First lateral tooth is roughly wedge-shaped, attenuated posteriorly, taller than the central tooth, with a subtriangular blade at the antero-dorsal edge. Major lateral tooth is thin, long, with a shaft broadened near the base. Head of the major lateral tooth is unicuspid, bent, claw-shaped, with an extended base ending with a long, sharp-pointed process directed inside. Inner small laterals with two spatula-shaped processes and prow-shaped outer small laterals.
A total of 22 gills on each side, extending from valve II to valve VII. The gut containing rusty brown particles of detritus.
Remarks
The new species is the third species within the genus Thermochiton and has a very similar morphology to T. undocostatus and T. papuaensis. The new species differs from T. undocostatus by the tegmentum of intermediate valves being covered by granules (vs. undulating costae in T. undocostatus) and having two types of marginal spicules: long, smooth spicules-needles and short, smooth blunt-topped spicules (vs. only long, smooth spicules in T. undocostatus). Thermochiton xui sp. nov. differs from T. papuaensis by having granules in the whole tegmentum of intermediate valves (vs. granules only in the jugal area and other areas with undulating costae in T. papuaensis); having two types of marginal spicules (vs. three types in T. papuaensis: long, smooth spicules and two kinds of ribbed spicules); and the first lateral tooth of the radula being approximately twice as long as the central tooth (vs. the first lateral tooth is approximately the same length as the central tooth in T. papuaensis).
Genetic Analysis
The ABGD analysis was performed on the COI gene sequences to determine whether the two specimens of T. xui sp. nov. were the same species and to conducted sequence-based species delimitation of the members of the family Ischnochitonidae (Figure 6). The ABGD analysis divided the 25 candidate species of chitons into reciprocal clades on the phylogenetic trees. The two specimens collected from the Haima cold seeps belonged to one species, T. xui sp. nov., which was consistent with the morphological identification. Additionally, both Stenoplax alata and Stenoplax purpurascens were respectively divided into two separate species. ML and BI analyses of the concatenated three-gene dataset facilitated the reconstruction of the phylogenetic tree, revealing similar topologies (Figure 7). The concatenated tree indicated that the new species was a sister group to most Ischnochiton taxa which is one of the four clades of the genus Ischnochiton. The species Ischnochiton rissoi, Ischnochiton tridentatus, and Ischnochiton hakodadensis were excluded from this main clade and formed three separate and distant clades. Likewise, the concatenated tree provided consistent support for the non-monophyly of the family Ischnochitonidae. The Lepidozona taxa formed a group but also contained Callistochiton jacobaeus and Ischnochiton hakodadensis. Furthermore, the outgroups were nested within the family Ischnochitonidae.
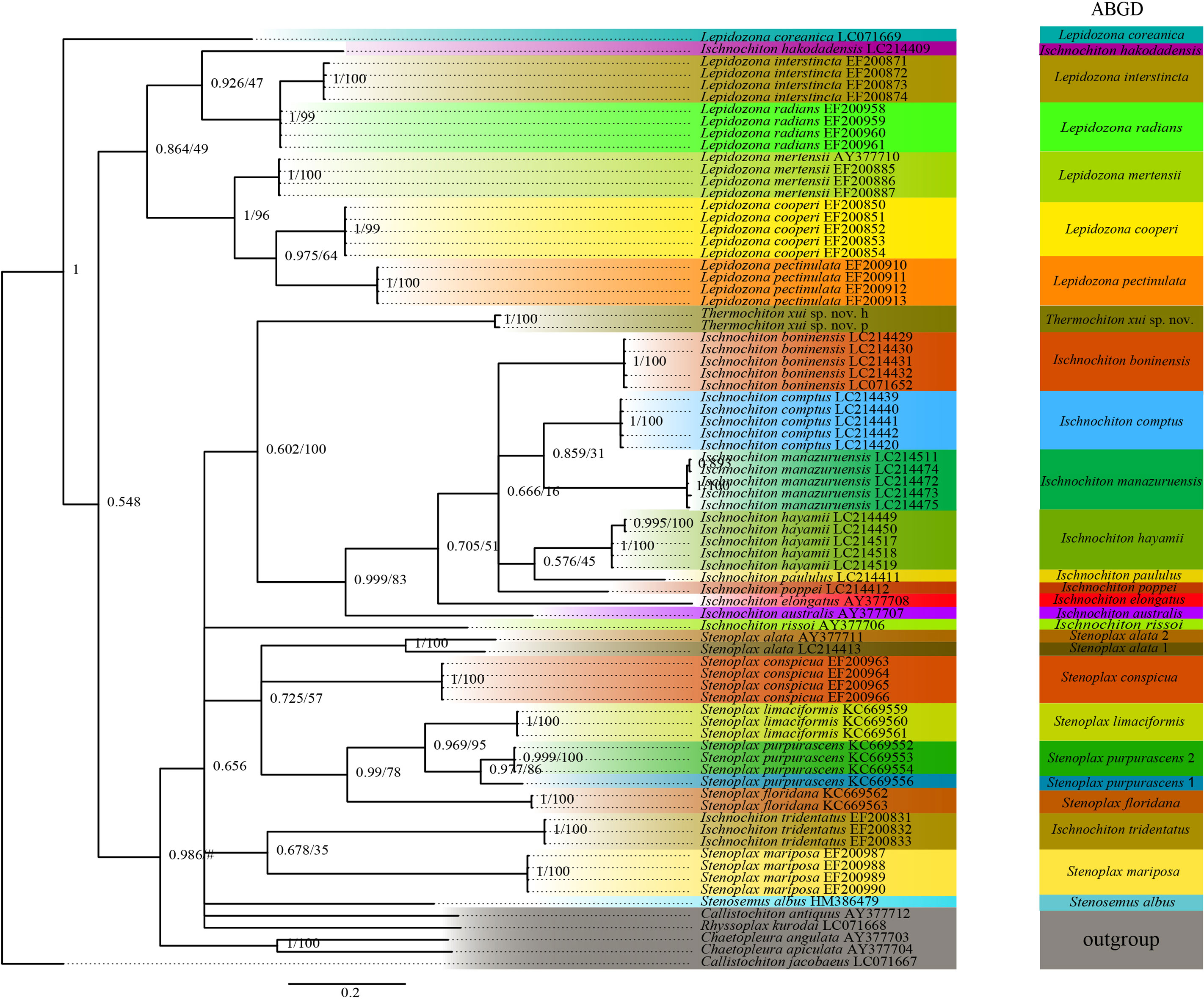
Figure 6 Phylogeny tree of the chitons based on the COI dataset. Numbers above branches refer to BI posterior probability (PP) and ML bootstrap scores (BS). The ABGD based on the COI alignment with the model Kimura (K80) TS/TV (2.0).
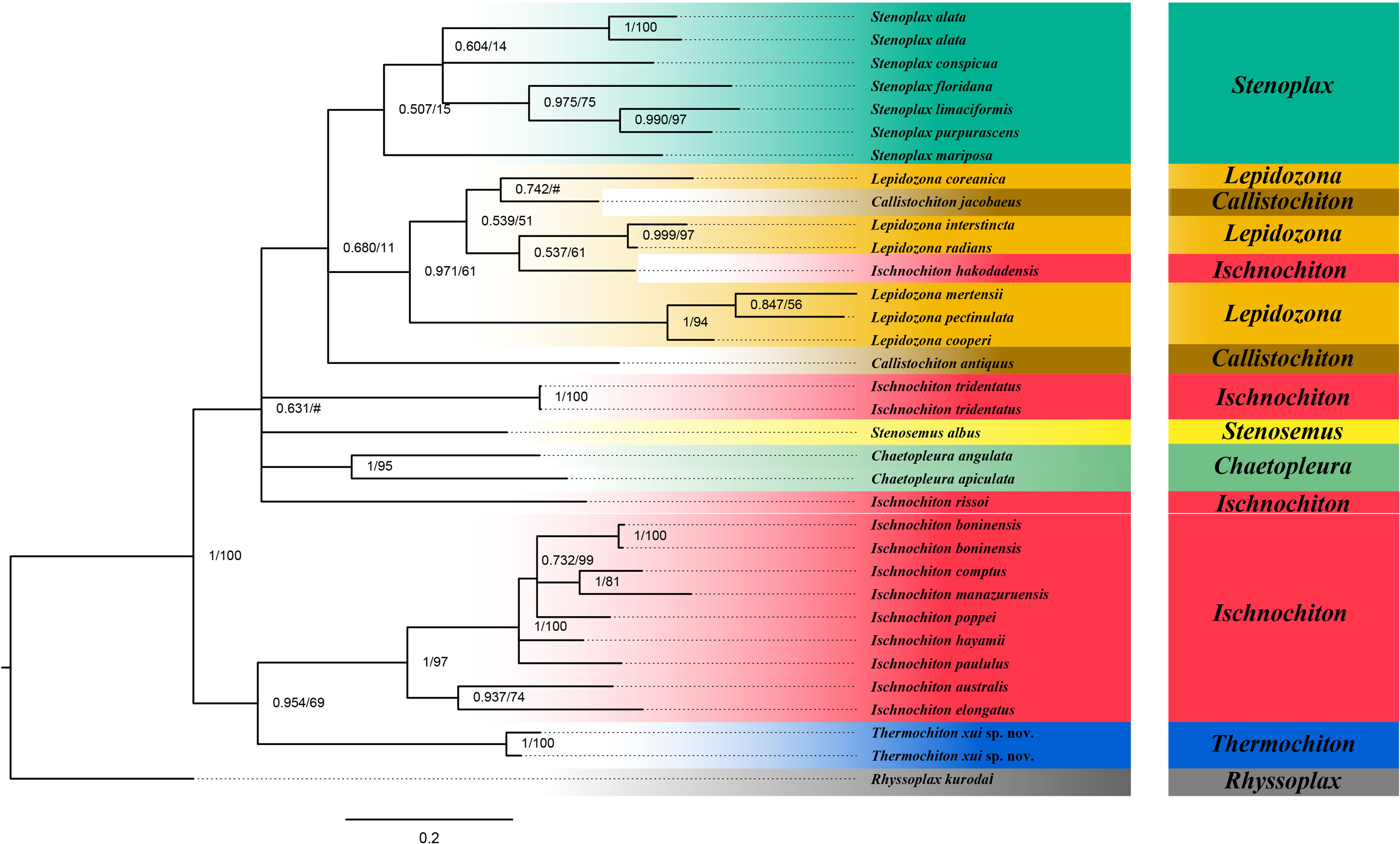
Figure 7 Phylogeny tree based on three genes concatenated sequences. Numbers above branches refer to BI posterior probability (PP) and ML bootstrap scores (BS).
The evolutionary tree produced using BEAST (Figure 8) shows the estimated divergence time for Ischnochitonidae species. According to the analysis of the log files using Tracer 1.7 software, all searches reached convergence, and all effective sample size (ESS) values were greater than 200. The calibrated estimate showed that T. xui sp. nov. was separated from most members of the genus Ischnochiton in the Late Miocene, roughly 7.31 MYA, with a 95% highest posterior density (HPD) interval of 5.10–10.07 million years. Radiation of the family Ischnochitonidae occurred mainly from the Late Miocene to Pliocene.
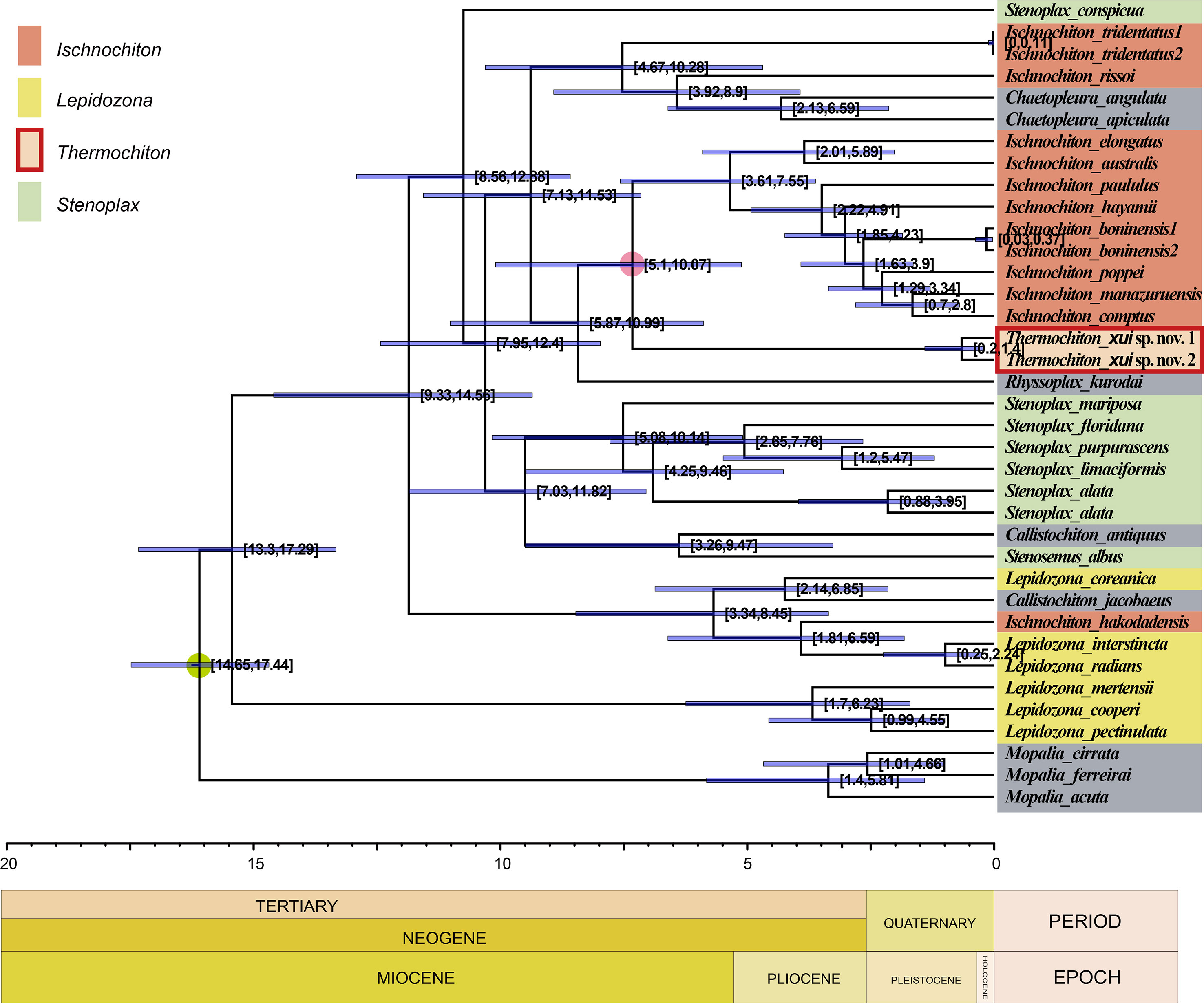
Figure 8 Chronogram of the family Ischnochitonidae obtained with an uncorrelated relaxed clock method (BEAST) on the combined data set (COI, 16S, and 28S genes). Divergent time estimates and 95% highest posterior probabilities were displayed on each node. The green dot indicates fossil calibration point. The red circle indicates split time of Thermochiton from its shallow water relatives.
Numerical Modeling
The numerical model in this study was designed to explore the evolutionary pathway of Thermochiton species and demonstrate how larvae in the three localities were connected by ocean currents. Thermochiton larvae were assumed to be passive floating particles in the OGCM to stimulate the spread process during PLD (Figure 9 and Supplementary Figures S1–S3). Even ignoring their time swimming from the bottom to the sea surface and assuming that larvae use the optimal way to spread, i.e., being carried by surface currents, larvae from Papua New Guinea needed at least 15 and 26 months to spread to the Okinawa Trough and Haima cold seeps, respectively (Supplementary Figure S1). Meanwhile, larvae from the Haima cold seeps required eight months to spread to the Okinawa Trough (Supplementary Figure S3). Notably, the model started in January when northeast (winter) monsoons dominated the South China Sea. Thus, the simulated northeastward surface current would be relatively slower during the first few months than that simulated by a model starting when southwest (summer) monsoons dominated the area. Nevertheless, the simulation ran for 36 months and did not reveal the possibility of the tracer diffusing from the Haima cold seeps or Okinawa Trough to Papua New Guinea (Supplementary Figures S3, S2).
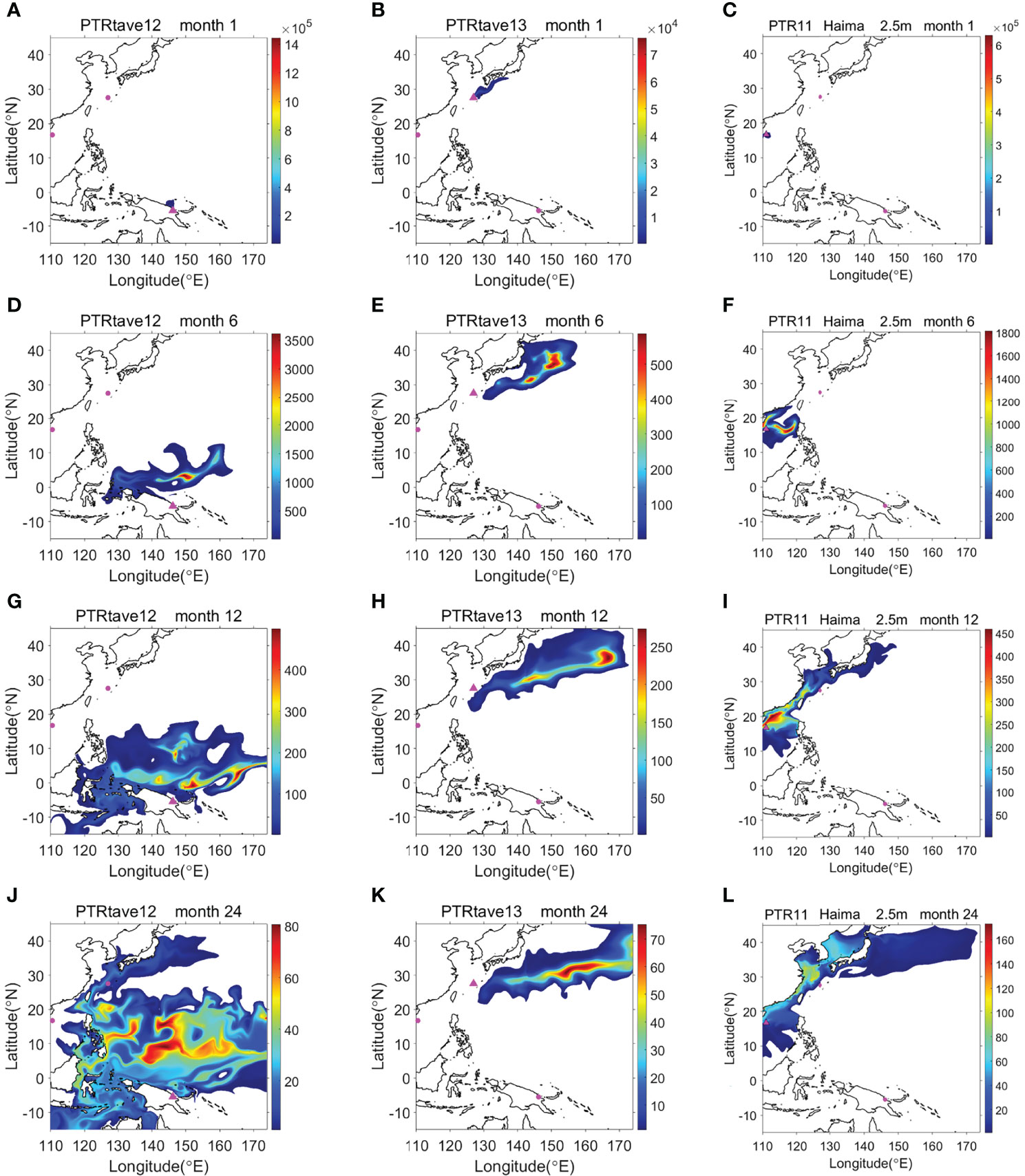
Figure 9 Dispersal path and relative content of chiton larvae released from Papua New Guinea (A, D, G, J), Okinawa Trough (B, E, H, K), and Haima cold seeps (C, F, I, L) as modeled by the OGCM for 1 month (A–C), 6 months (D–F), 12 months (G–I) and 24 months (J–L).
Although development information about deep-sea chitons is lacking, larvae of shallow-water chitons are mostly lecithotrophic. As a result, deep-sea chitons are more likely spread to the seafloor or deep ocean layer than to float to the surface. Further, the current velocity at the bottom or in the deep ocean layer is much slower than that at the surface, suggesting that deep-sea chitons would need much longer to spread than those floating at the surface.
Discussion
The discovery of T. xui sp. nov. increased the number of known members of the genus Thermochiton. Currently, the three species of the genus likely all live in areas of chemosynthetic activity. The genus Thermochiton was established for T. undocostatus Saito & Okutani, 1990, a chiton found in hydrothermal vents at the central Okinawa Trough, East China Sea (Saito and Okutani, 1990). The second member of the genus, T. papuaensis, was reported near Papua New Guinea and is likely to inhabit communities with chemical activity (Sirenko, 2020). Our new species was sampled from the Haima cold seeps in the South China Sea. The distribution areas of the genus are geographically far removed, located in the southwest and northwest Pacific. The type locality of T. papuaensis is approximately 4,637 km away from this sampling site and approximately 4,217 km away from the type locality of T. undocostatus (Figure 1).
The new species is the only species with molecular information in the genus. It is impossible to analyze the phylogenetic relationship between the three Thermochiton species or infer how their ancestors spread based merely on morphological or molecular information. The modeling results in this study indicated that one Thermochiton species is unlikely to have crossed the complex Pacific western boundary currents to reach the three localities in which the genus has been reported. The western Pacific vents in distant basins are potentially connected by the Kuroshio Current (Mitarai et al., 2016). In addition, the South Equatorial Current can distribute vent species more than 4,000 km within the southwest Pacific. Nevertheless, the results of our numerical model suggested that at least 15 months were required for larvae to spread from Papua New Guinea to the Okinawa Trough via surface currents. Northward evolutionary migration is highly more likely than southward migration for Pacific deep-sea chitons. The upper prediction of the model reported by Yearsley and Sigwart (2011) estimated no more than 240 days (i.e., 8 months) for the PLD of deep-sea chitons. The deep-sea mollusk with the longest PLD is the cold seep mussel B. naticoidea, whose larvae reportedly disperse for 7–12 months at the surface before settling (Arellano et al., 2014). The larvae of the seastar Mediaster aequalis can reportedly persist for approximately 13 months before settling (Birkeland et al., 1971). Nevertheless, it was estimated that even the longest PLD could not be expected to exceed 400 days (O’Connor et al., 2007). Thus, one Thermochiton species is unlikely to have a sufficiently long PLD and dispersal ability to cross the complex Pacific western boundary currents, even under ideal conditions and with extremely strong surface currents. It is more likely that currents carried the pelagic larvae of their ancestors to spread and radiate into different species along the evolutionary pathway during colonization. We hypothesize that (1) ‘stepping-stones’ facilitated their colonization, i.e., suitable habitats for chitons between the southwest and northwest Pacific, such as seeps, vents, sunken wood, or carcasses, and/or (2) evolutionary ‘bridge species’ existed during their spread, i.e., chitons distributed at ‘stepping-stone’ sites that have not yet been found or became extinct with the disappearance of these ephemeral habitats. Therefore, additional expeditions are needed to further explore the deep-sea fauna and collect more data in order to reconstruct the evolutionary history of this genus.
Most deep-water chitons belong to the order Lepidopleurida, a basal clade of the class Polyplacophora. Sigwart (2017) suggested that this clade originated at a depth of approximately 500 m and radiated to either deep-sea or coastal areas. Most of the radiation to sunken wood occurred in the Jurassic. Many lepidopleuran chitons diversified during the Cenozoic and invaded the deep sea and different chemosynthetic habitats. Many other mollusks in hydrothermal vents, seeps, and organic falls have the pattern of Cenozoic radiation in the deep sea (Johnson et al., 2010; Lorion et al., 2013; Taylor et al., 2014). Indeed, our molecular clock analysis revealed that the genus Thermochiton originated from a shallow-sea ancestor around 5.10 to 10.07 MYA in the Late Miocene. The genus follows the “onshore to offshore” evolutionary pattern (Jablonski et al., 1983; Vrijenhoek, 2013), but occurred independently of the invasion of lepidopleuran chitons into chemosynthetic habitats.
A total of 23 species of deep-sea chitons have been identified in the bathyal and abyssal zones of the East and South China Seas to date (Kaas, 1979; Saito and Okutani, 1990; Xu, 1990; Saito, 2005; Saito, 2011; Saito, 2013; Sirenko, 2016b; Sigwart and Chen, 2018; Sirenko, 2018). Among them, 15 species live in hydrothermal vents, cold seeps, or sunken wood habitats (Table 2). Chemosynthetic areas are the main habitats of deep-sea chitons. Sirenko (2018) discovered 16 chiton species off Taiwan, 13 of which were new occurrences and two were new species, based on two exploration missions in 2000 and 2001. These data significantly increased the deep-sea chiton biodiversity in the East and South China Seas. To date, only four species of deep-sea chitons have been found in the South China Sea (Leptochiton vietnamensis Sirenko, 1998; Parachiton laevisquamatus Sirenko, 2015; Loricella. scissurata (Xu, 1990); Lepidozona acostata Sirenko, 2016; and Thermochiton xui sp. nov.). Sirenko and Zhang (2019) reported 30 chiton species from the vicinity of Hainan Island. However, the deep sea and numerous islands of the South China Sea have not been adequately sampled. The number of chitons currently recorded in the deep and shallow waters of the South China Sea is far from exhaustive compared with the nearly 100 species of chiton recorded in Japan (Slieker, 2000; Saito, 2001b; Saito, 2017) and the 68 species of chiton found in Vietnam (Sirenko, 2016b). The abundance of chiton species found in other locations suggests that the South China Sea has a high degree of biodiversity waiting to be discovered.
Our phylogenetic analysis based on the concatenated sequences from three gene regions supported the non-monophyly of the family Ischnochitonidae, as well as that of the genera Ischnochiton, Lepidozona, and Callistochiton (Figure 7). The outgroups were nested within the clade of the family Ischnochitonidae, similar to the findings reported by Owada (2018). The results suggest that the molecular data are not consistent with the assignment of species to genera and families based on plesiomorphic similarities. Both morphologic and genetic data indicated the complexity of the family Ischnochitonidae. Members of the genus Connexochiton Kaas, 1979 resemble those of the genus Thermochiton. Members of both genera have rounded granules at the distal end of the dorsal girdle scales, and the heads of the major lateral teeth of the radulae are unicuspidal, bent, and sickle-shaped. However, members of the genus Thermochiton have pores of aesthetes distributed throughout the tegmentum with concentric, undulating costae, whereas the pores of aesthetes of members of the genus Connexochiton are only distributed in granules. The genus Subterenochiton Iredale & Hull, 1924 is similar to members of the genus Thermochiton but differs by the presence of flattened, closely set, and same-sized tegmentum granules, as well as a single slit per side in the intermediate valves. The genus Thermochiton can be clearly distinguished from most Ischnochiton species by the following characteristics: small body size vs. medium to large body size; tegmentum granules raised vs. flat granules; intermediate valves with 2 slits per side vs. 1 slit per side; dorsal scales with granules vs. no granules; head of the major lateral tooth of the radula is unicuspidal vs. bicuspidal. Meanwhile, Ischnochiton luteoroseus Suter, 1907, Ischnochiton granulifer Thiele, 1909, and Ischnochiton albinus Thiele, 1911 also share similarities with members of the genus Thermochiton, including rounded granules on the dorsal girdle scales, unicuspidal head of the major lateral tooth of the radula, and a distinct protuberance on the major lateral tooth head. Interestingly, several species of the genus Lepidozona, Ischnochiton, Subterenochiton, and Callistochiton have dorsal scales with granules and/or longitudinal ribs, which is similar to members of the genus Thermochiton. Thus, these taxa need to be re-examined and more taxon sampling is needed. The current classification system of chitons was established based mainly on the morphology of valves, spicules, and girdle processes. Few taxonomic studies have combined morphology with molecular biology (Sigwart et al., 2010; Irisarri et al., 2020). However, as additional molecular data become available, classification uncertainty will undoubtedly arise. Emerging new data will provide insights into phylogenetic relationships and improve the stepwise systematics approach. For example, the previous morphological classification placed Ischnochiton. hakodadensis in the genus Ischnochiton, but molecular analysis revealed that the species was nested within the genus Lepidozona [(present study and findings reported by Owada (2018)]. The current information is insufficient to suggest any rearrangement of the family Ischnochitonidae; thus, this family remains poorly defined. Integrative taxonomy incorporating molecular data and morphological evidence will help solve this problem (Zhang et al., 2020). More evidence is needed to clarify phylogenetic relationships and improve stepwise systematics of chitons.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
JZ conceived and designed this project. HW, JZ, and BS performed morphological examination and description. HL and JZ conducted molecular analyses. XW and CL preformed the numerical modeling. DD and XL collected the samples. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This work was supported by the Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) (No. 2022QNLM050102-2), the National Key Research and Development Program of China (2021YFE0193700), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB42000000, and XDA22050203), the National Natural Science Foundation of China (31772422, and 42176114), the International Science Partnership Program of the Chinese Academy of Sciences (No. 133137KYSB20200002), and the Senior User Project of RV KEXUE (KEXUE2020GZ01).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are grateful to Editor Prof. Jian-Wen Qiu (Hong Kong Baptist University), Prof. Douglas Eernisse (California State University Fullerton), and an Dr. Julia Sigwart (Queen’s University Belfast, United Kingdom), for comments and suggestions on the manuscript. Thanks also to Ms. Shanshan Wang (Institute of Oceanology, Chinese Academy of Sciences) for her assistance with SEM procedures and the photos.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.889022/full#supplementary-material
References
Adams D. K., Arellano S. M., Govenar B. (2012). Larval Dispersal: Vent Life in the Water Column. Oceanography 25 (1), 256–268. doi: 10.5670/oceanog.2012.24
Arellano S. M., Van Gaest A. L., Johnson S. B., Vrijenhoek R. C., Young C. M. (2014). Larvae From Deep-Sea Methane Seeps Disperse in Surface Waters. Proc. R. Soc B: Biol. Sci. 281 (1786), 20133276. doi: 10.1098/rspb.2013.3276
Atlas R., Hoffman R. N., Ardizzone J., Leidner S. M., Jusem J. C., Smith D. K., et al. (2011). A Cross-Calibrated, Multiplatform Ocean Surface Wind Velocity Product for Meteorological and Oceanographic Applications. Bull. Amer. Meteor. Soc 92 (2), 157–174. doi: 10.1175/2010bams2946.1
Beedessee G., Watanabe H., Ogura T., Nemoto S., Yahagi T., Nakagawa S., et al. (2013). High Connectivity of Animal Populations in Deep-Sea Hydrothermal Vent Fields in the Central Indian Ridge Relevant to Its Geological Setting. PloS One 8 (12), e81570. doi: 10.1371/journal.pone.0081570
Birkeland C., Chia F.-S., Strathmann R. R. (1971). Development, Substratum Selection, Delay of Metamorphosis and Growth in Seastar, Mediaster Aequalis Stimpson. Biol. Bull. 141 (1), 99–108. doi: 10.2307/1539994
Bouchet P., Warén A. (1994). “Ontogenetic Migration and Dispersal of Deep-Sea Gastropod Larvae,” in Reproduction, Larval Biology and Recruitment in the Deep-Sea Benthos. Eds. Young C. M., Eckelbarger K. J. (New York: Columbia University Press), 98–117.
Chen C., Okutani T., Liang Q., Qiu J. W. (2018). A Noteworthy New Species of the Family Vesicomyidae From the South China Sea (Bivalvia: Glossoidea). Venus 76 (1-4), 29–37. doi: 10.18941/venus.76.1-4_29
Darriba D., Taboada G. L., Doallo R., Posada D. (2012). Jmodeltest 2: More Models, New Heuristics and Parallel Computing. Nat. Methods 9 (8), 772–772. doi: 10.1038/nmeth.2109
Dong D., Li X., Yang M., Gong L., Li Y., Sui J., et al. (2021). Report of Epibenthic Macrofauna Found From Haima Cold Seeps and Adjacent Deep-Sea Habitats, South China Sea. Mar. Life Sci. Technol. 3 (1), 1–12. doi: 10.1007/s42995-020-00073-9
Fukumori H., Kano Y. (2014). Evolutionary Ecology of Settlement Size in Planktotrophic Neritimorph Gastropods. Mar. Biol. 161 (1), 213–227. doi: 10.1007/s00227-013-2330-5
García-Ríos C. I., Pérez-Pérez N. M., Fernández-López J., Fuentes F. A. (2014). Calibrating the Chitons (Mollusca: Polyplacophora) Molecular Clock With the Mitochondrial DNA Cytochrome C Oxidase I Gene. Rev. Biol. Mar. Oceanogr. 49 (2), 193–207. doi: 10.4067/s0718-19572014000200003
Gent P. R., McWilliams J. C. (1990). Isopycnal Mixing in Ocean Circulation Models. J. Phys. Oceanogr. 20 (1), 150–155. doi: 10.1175/1520-0485(1990)020<0150:imiocm>2.0.co;2
Irisarri I., Uribe J. E., Eernisse D. J., Zardoya R. (2020). A Mitogenomic Phylogeny of Chitons (Mollusca: Polyplacophora). BMC Evol. Biol. 20 (1), 22. doi: 10.1186/s12862-019-1573-2
Jablonski D., Sepkoski J. J., Bottjer D. J., Sheehan P. M. (1983). Onshore-Offshore Patterns in the Evolution of Phanerozoic Shelf Communities. Science 222 (4628), 1123–1125. doi: 10.1126/science.222.4628.1123
Jiang J., Huang Y., Liang Q., Zhang J. (2019). Description of Two New Species (Bivalvia: Vesicomyidae, Verticordiidae) From a Cold Seep in the South China Sea. Nautilus 133 (3-4), 94–101.
Johnson S. B., Warén A., Lee R. W., Kano Y., Kaim A., Davis A., et al. (2010). Rubyspira, New Genus and Two New Species of Bone-Eating Deep-Sea Snails With Ancient Habits. Biol. Bull. 219 (2), 166–177. doi: 10.1086/BBLv219n2p166
Kaas P. (1979). On a Collection of Polyplacophora (Mollusca, Amphineura) From the Bay of Biscay. Bull. Mus. Natl. Hist. Nat. Sect. A Zool. 4 (1), 1–13.
Kaas P., Van Belle R. A. (1980). Catalogue of Living Chitons (Mollusca: Polyplacophora) (Rotterdam: W. Backhuys).
Kaas P., Van Belle R. A. (1987). Monograph of Living Chitons (Mollusca: Polyplacophora), Volume 3 Ischnochitonidae: Chaetopleurinae and Ischnochitoninae - Pars. Addition to Vols 1 & 2 (Leiden: E. J. Brill).
Katoh K., Standley D. M. (2013). MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 30 (4), 772–780. doi: 10.1093/molbev/mst010
Kelly R. P., Eernisse D. J. (2007). Southern Hospitality: A Latitudinal Gradient in Gene Flow in the Marine Environment. Evolution 61 (3), 700–707. doi: 10.1111/j.1558-5646.2007.00055.x
Kelly R. P., Eernisse D. J. (2008). Reconstructing a Radiation: The Chiton Genus Mopalia in the North Pacific. Invertebr. Syst. 22 (1), 17–28. doi: 10.1071/is06021
Kimura M. (1980). A Simple Method for Estimating Evolutionary Rates of Base Substitutions Through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 16 (2), 111–120. doi: 10.1007/BF01731581
Large W. G., McWilliams J. C., Doney S. C. (1994). Oceanic Vertical Mixing: A Review and a Model With a Nonlocal Boundary Layer Parameterization. Rev. Geophys. 32 (4), 363. doi: 10.1029/94rg01872
Layton K. K., Martel A. L., Hebert P. D. (2014). Patterns of DNA Barcode Variation in Canadian Marine Molluscs. PloS One 9 (4), e95003. doi: 10.1371/journal.pone.0095003
Liang Q., Hu Y., Feng D., Peckmann J., Chen L., Yang S., et al. (2017). Authigenic Carbonates From Newly Discovered Active Cold Seeps on the Northwestern Slope of the South China Sea: Constraints on Fluid Sources, Formation Environments, and Seepage Dynamics. Deep-Sea Res. Part I: Oceanogr. Res. Pap., 124, 31–41. doi: 10.1016/j.dsr.2017.04.015
Lorion J., Kiel S., Faure B., Kawato M., Ho S. Y. W., Marshall B., et al. (2013). Adaptive Radiation of Chemosymbiotic Deep-Sea Mussels. Proc. R. Soc B: Biol. Sci. 280 (1770), 20131243. doi: 10.1098/rspb.2013.1243
Marshall J., Hill C., Perelman L., Adcroft A. (1997). Hydrostatic, Quasi-Hydrostatic, and Nonhydrostatic Ocean Modeling. J. Geophys. Res. Oceans 102 (C3), 5733–5752. doi: 10.1029/96jc02776
Mitarai S., Watanabe H., Nakajima Y., Shchepetkin A. F., McWilliams J. C. (2016). Quantifying Dispersal From Hydrothermal Vent Fields in the Western Pacific Ocean. Proc. Natl. Acad. Sci. 113 (11), 2976–2981. doi: 10.1073/pnas.1518395113
O’Connor M. I., Bruno J. F., Gaines S. D., Halpern B. S., Lester S. E., Kinlan B. P., et al. (2007). Temperature Control of Larval Dispersal and the Implications for Marine Ecology, Evolution, and Conservation. Proc. Natl. Acad. Sci. 104 (4), 1266–1271. doi: 10.1073/pnas.0603422104
Okusu A., Schwabe E., Eernisse D. J., Giribet G. (2003). Towards a Phylogeny of Chitons (Mollusca, Polyplacophora) Based on Combined Analysis of Five Molecular Loci. Org. Div. Evol. 3 (4), 281–302. doi: 10.1078/1439-6092-00085
Owada M. (2016). A New Cryptic Species Distinguished From Ischnochiton Comptus (Gould 1859) (Polyplacophora: Ischnochitonidae) in Central Honshu, Japan. Molluscan Res. 36 (4), 255–263. doi: 10.1080/13235818.2016.1150772
Owada M. (2018). Phylogenetic Relationships Among Japanese Species of the Genus Ischnochiton (Polyplacophora: Ischnochitonidae), Including a New Species. Zool. Sci. 35 (3), 281–291. doi: 10.2108/zs170106
Puillandre N., Lambert A., Brouillet S., Achaz G. (2012). ABGD, Automatic Barcode Gap Discovery for Primary Species Delimitation. Mol. Ecol. 21 (8), 1864–1877. doi: 10.1111/j.1365-294X.2011.05239.x
Redi M. H. (1982). Oceanic Isopycnal Mixing by Coordinate Rotation. J. Phys. Oceanogr. 12 (10), 1154–1158. doi: 10.1175/1520-0485(1982)012<1154:OIMBCR>2.0.CO;2
Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., et al. (2012). MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 61 (3), 539–542. doi: 10.1093/sysbio/sys029
Saito H. (1997). Deep-Sea Chiton Fauna of Suruga Bay (Mollusca: Polyplacophora) With Descriptions of Six New Species. Nat. Sci. Mus. Monogr. 12, 31–58.
Saito H. (2001a). Chitons (Mollusca: Polyplacophora) Collected by the R/V Kotaka-Maru From Tosa Bay, Western Japan, With Descriptions of Two New Species. Nat. Sci. Mus. Monogr. 20, 101–119.
Saito H. (2001b). Shallow-Water Chitons (Mollusca: Polyplacophora) of Hachijo Island, Izu Islands, Japan. Mem. Natn. Sci. Mus. 37, 193–202.
Saito H. (2004). Phylogenetic Significance of the Radula in Chitons, With Special Reference to the Cryptoplacoidea (Mollusca: Polyplacophora). Boll. Malacol. 39(Supplement 5), 83–104.
Saito H. (2005). Shelf and Bathyal Chitons (Mollusca: Polyplacophora) From the Nansei Islands, Southwestern Japan. Nat. Sci. Mus. Monogr. 29, 101–113.
Saito H. (2011). Chitons (Mollusca: Polyplacophora) From Submarine Banks Off Izu Islands and Bōsō Peninsula, Japan. Mem. Natn. Mus. Nat. Sci. Tokyo 47, 65–81.
Saito H. (2012). Second Specimen of a Rare Deep-Sea Chiton, Deshayesiella Sinica (Xu 1990) (Polyplacophora, Lepidopleurida, Protochitonidae) From Northern Japan. Bull. Natl. Mus. Nat. Sci. 38, 7–11.
Saito H. (2013). A New Species of Lepidozona (Mollusca, Polyplacophora, Ischnochitonidae) From Okinawa Trough, East China Sea. Bull. Natl. Mus. Nat. Sci. 39, 5–10.
Saito H. (2017). “Class Polyplacophora,” in Marine Mollusks in Japan, 2nd ed. Ed. Okutani T. (Tokyo: Tokai University Press), 728–738.
Saito H., Fujikura K., Tsuchida S. (2008). Chitons (Mollusca: Polyplacophora) Associated With Hydrothermal Vents and Methane Seeps Around Japan, With Descriptions of Three New Species. Am. Malacol. Bull. 25 (1), 113–124. doi: 10.4003/0740-2783-25.1.113
Saito H., Okutani T. (1990). Two New Chitons (Mollusca: Polyplacophora) From a Hydrothermal Vent Site of the Iheya Small Ridge, Okinawa Trough, East China Sea. Venus 49 (3), 165–179. doi: 10.18941/venusjjm.49.3_165
Schwabe E. (2006). Chitons (Mollusca, Polyplacophora) Collected During the Thai-Danish Bioshelf Surveys, (1996-2000) in the Andaman Sea, Indian Ocean. J. Zool. Soc. Wallacea 2, 19–28.
Schwabe E., Sellanes J. (2010). Revision of Chilean Bathyal Chitons (Mollusca: Polyplacophora) Associated With Cold-Seeps, Including Description of a New Species of Leptochiton (Leptochitonidae). Org. Div. Evol. 10 (1), 31–55. doi: 10.1007/s13127-009-0002-6
Sigwart J. D. (2017). Deep Trees: Woodfall Biodiversity Dynamics in Present and Past Oceans. Deep-Sea Res. II: Top. Stud. Oceanogr. 137, 282–287. doi: 10.1016/j.dsr2.2016.06.021
Sigwart J. D., Chen C. (2018). A New Deep Water Chiton (Mollusca: Polyplacophora) From Hydrothermal Vent Ecosystems in the Okinawa Trough, Japan. Zootaxa 4531 (3), 430–436. doi: 10.11646/zootaxa.4531.3.7
Sigwart J. D., Schwabe E., Saito H., Samadi S., Giribet G. (2010). Evolution in the Deep Sea: A Combined Analysis of the Earliest Diverging Living Chitons (Mollusca: Polyplacophora: Lepidopleurida). Invertebr. Syst. 24 (6), 560–572. doi: 10.1071/is10028
Sigwart J. D., Sirenko B. I. (2012). Deep-Sea Chitons From Sunken Wood in the West Pacific (Mollusca: Polyplacophora: Lepidopleurida): Taxonomy, Distribution, and Seven New Species. Zootaxa 9 (3195), 1–38. doi: 10.11646/zootaxa.3195.1.1
Sirenko B. I. (1988). A New Genus of Deep Sea Chitons Ferreiraella Gen. N. (Lepidopleurida, Leptochitonidae) With a Description of a New Ultra-Abyssal Species. Zool. Zhurnal 67 (12), 1776–1786.
Sirenko B. I. (1998). One More Deep Water Chiton Leptochiton Vietnamensis Sp. N. (Mollusca, Polyplacophora) Living and Feeding on Sunken Wood From the South China Sea. Ruthenica 8 (1), 1–6.
Sirenko B. I. (2001). “Deep-Sea Chitons (Mollusca, Polyplacophora) From Sunken Wood Off New Caledonia and Vanuatu,” in Tropical Deep-Sea Benthos, vol. 22 . Eds. Bouchet P., Marshall B. (France: Muséum National d’Histoire Naturelle), 39–71. doi: 10.18941/venus.65.1-2_27
Sirenko B. I. (2006). New Outlook on the System of Chitons (Mollusca: Polyplacophora). Venus 65 (1-2), 27–49.
Sirenko B. I. (2015). New Species of Parachiton (Mollusca: Polyplacophora) From the South China Sea. Zoosystematica Ross. 24 (2), 143–147. doi: 10.31610/zsr/2015.24.2.143
Sirenko B. I. (2016a). New and Rare Species of the Genus Lepidozona (Mollusca: Polyplacophora) From the South China, East China and the Philippine Seas. Bull. Russ. Far East Malacol. Soc 20 (2), 28–46.
Sirenko B. I. (2016b). “Seven-Year Taxonomical Investigation of Chitons in Vietnam,” in in Proceedings of the Conference “Developing Life-Supporting Marine Ecosystems Along With the Asia-Pacific Coast a Synthesis of Physical and Biological Data for the Science-Based Management and Socio-Ecological Policy Making”. Ed. Dautova T. N. (Vladivostok-Nha Trang: Dalnauka), 82–86.
Sirenko B. I. (2018). Taiwanese Deep-Water Chitons (Mollusca: Polyplacophora) and Survey of Chiton Fauna of Taiwan. Zootaxa 4422 (3), 301–344. doi: 10.11646/zootaxa.4422.3.1
Sirenko B. I. (2020). A Second Species of the Genus Thermochiton Saito Et Okutani 1990 (Mollusca: Polyplacophora). Ruthenica 30 (1), 7–12. doi: 10.35885/ruthenica.2021.30(1).2
Sirenko B. I. (2021). Composition of the Genus Loricella (Mollusca: Polyplacophora: Loricidae) and the Description of Two New Species. Zootaxa 4981 (2), 275–300. doi: 10.11646/zootaxa.4981.2.4
Sirenko B. I., Sellanes J. (2016). Update of the Genus Leptochiton (Mollusca: Polyplacophora) in Chilean Deep Waters: Three New Reports and Description of Two New Species. Zootaxa 4173 (3), 259–283. doi: 10.11646/zootaxa.4173.3.5
Sirenko B. I., Zhang J. (2019). Chitons (Mollusca: Polyplacophora) of Hainan Island and Vicinity, South China Sea. Zootaxa 4564 (1), 1–40. doi: 10.11646/zootaxa.4564.1.1
Slieker F. J. A. (2000). Chitons of the World: An Illustrated Synopsis of Recent Polyplacophora (Italy: Mostra mondiale malacologia, Cupra Marittima).
Stamatakis A. (2014). RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 30 (9), 1312–1313. doi: 10.1093/bioinformatics/btu033
Suchard M. A., Lemey P., Baele G., Ayres D. L., Drummond A. J., Rambaut A. (2018). Bayesian Phylogenetic and Phylodynamic Data Integration Using BEAST 1.10. Virus Evol. 4 (1), vey016. doi: 10.1093/ve/vey016
Sun Y., Liang Q., Sun J., Yang Y., Tao J., Liang J., et al. (2018). The Mitochondrial Genome of the Deep-Sea Tubeworm Paraescarpia Echinospica (Siboglinidae, Annelida) and its Phylogenetic Implications. Mitochondrial DNA B: Res. 3 (1), 131–132. doi: 10.1080/23802359.2018.1424576
Taylor J. D., Glover E. A., Williams S. T. (2014). Diversification of Chemosymbiotic Bivalves: Origins and Relationships of Deeper Water Lucinidae. Biol. J. Linn. Soc 111 (2), 401–420. doi: 10.1111/bij.12208
Teixeira S., Cambon-Bonavita M.-A., Serrão E. A., Desbruyéres D., Arnaud-Haond S. (2011). Recent Population Expansion and Connectivity in the Hydrothermal Shrimp Rimicaris Exoculata Along the Mid-Atlantic Ridge. J. Biogeogr. 38 (3), 564–574. doi: 10.1111/j.13652699.2010.02408.x
Tyler P. A., Young C. M. (1999). Reproduction and Dispersal at Vents and Cold Seeps. J. Mar. Biol. Assoc. U. K. 79 (2), 193–208. doi: 10.1017/s0025315499000235
Tyler P. A., Young C. M. (2003). Dispersal at Hydrothermal Vents: A Summary of Recent Progress. Hydrobiologia 503 (1-3), 9–19. doi: 10.1023/b:hydr.0000008492.53394.6b
Vaidya G., Lohman D. J., Meier R. (2011). SequenceMatrix: Concatenation Software for the Fast Assembly of Multi-Gene Datasets With Character Set and Codon Information. Cladistics 27 (2), 171–180. doi: 10.1111/j.1096-0031.2010.00329.x
Vrijenhoek R. C. (2013). On the Instability and Evolutionary Age of Deep-Sea Chemosynthetic Communities. Deep-Sea Res. II - Top. Stud. Oceanogr. 92, 189–200. doi: 10.1016/j.dsr2.2012.12.004
Warén A., Bouchet P. (2001). Gastropoda and Monoplacophora From Hydrothermal Vents and Seeps: New Taxa and Records. Veliger 44, 116–231.
Wu S. K., Okutani T. (1984). The Deepsea Chitons (Mollusca: Polyplacophora) Collected by the R/V Soyo-Maru From Japan. I, Lepidopleuridae. Venus 44 (1), 1–31. doi: 10.18941/venusjjm.43.1_1
Xu F. (1990). New Genus and Species of Polyplacophora (Mollusca) From the East China Sea. Chin. J. Oceanol. Limnol. 8 (4), 374–377. doi: 10.1007/BF02849683
Xu T., Feng D., Tao J., Qiu J.-W. (2019). A New Species of Deep-Sea Mussel (Bivalvia: Mytilidae: Gigantidas) From the South China Sea: Morphology, Phylogenetic Position, and Gill-Associated Microbes. Deep-Sea Res. I: Oceanogr. Res. Pap. 146, 79–90. doi: 10.1016/j.dsr.2019.03.001
Yahagi T., Fukumori H., Warén A., Kano Y. (2019). Population Connectivity of Hydrothermal-Vent Limpets Along the Northern Mid-Atlantic Ridge (Gastropoda: Neritimorpha: Phenacolepadidae). J. Mar. Biol. Assoc. U. K. 99 (1), 179–185. doi: 10.1017/s0025315417001898
Yahagi T., Kayama Watanabe H., Kojima S., Kano Y. (2017). Do Larvae From Deep-Sea Hydrothermal Vents Disperse in Surface Waters? Ecology 98 (6), 1524–1534. doi: 10.1002/ecy.1800
Yearsley J. M., Sigwart J. D. (2011). Larval Transport Modeling of Deep-Sea Invertebrates can Aid the Search for Undiscovered Populations. PLoS One 6 (8), e23063. doi: 10.1371/journal.pone.0023063
Zhang S., Zhang S. (2017). Description of Pyropelta Elongata Sp. Nov. (Gastropoda, Pyropeltidae) From a Methane Seep Area in the South China Sea. Am. Malacol. Bull. 35 (1), 51–54. doi: 10.4003/006.035.0106
Zhang S., Zhang J., Zhang S. (2016). A New Species of Bathyacmaea (Gastropoda: Pectinodontidae) From a Methane Seep Area in the South China Sea. Nautilus 130 (1), 1–4.
Zhang S., Zhang J., Zhang S. (2020). Integrative Taxonomy Reveals New Taxa of Trochidae (Gastropoda: Vetigastropoda) From Seamounts in the Tropical Western Pacific. Deep-Sea Res. I: Oceanogr. Res. Pap. 159, 103234. doi: 10.1016/j.dsr.2020.103234
Keywords: chiton, Thermochiton, Ischnochitonidae, Haima cold seeps, South China Sea, deep-sea, chemosynthetic habitat, numerical model
Citation: Wang H, Liu H, Wang X, Zhang J, Sirenko BI, Liu C, Dong D and Li X (2022) Stirring the Deep, Disentangling the Complexity: Report on the Third Species of Thermochiton (Mollusca: Polyplacophora) From Haima Cold Seeps. Front. Mar. Sci. 9:889022. doi: 10.3389/fmars.2022.889022
Received: 03 March 2022; Accepted: 13 June 2022;
Published: 18 July 2022.
Edited by:
Jian-Wen Qiu, Hong Kong Baptist University, Hong Kong SAR, ChinaReviewed by:
Douglas Eernisse, California State University, Fullerton, United StatesJulia Sigwart, Queen’s University Belfast, United Kingdom
Copyright © 2022 Wang, Liu, Wang, Zhang, Sirenko, Liu, Dong and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junlong Zhang, zhangjl@qdio.ac.cn
†These authors have contributed equally to this work
 Hao Wang1,2†
Hao Wang1,2†  Junlong Zhang
Junlong Zhang Chuanyu Liu
Chuanyu Liu Dong Dong
Dong Dong Xinzheng Li
Xinzheng Li