Morphology and molecular phylogeny of two new Aspidisca species (Ciliophora, Spirotrichea, Euplotida) collected from subtropical coastal waters in China
- 1School of Marine Sciences, Ningbo University, Ningbo, China
- 2Laboratory of Marine Protozoan Biodiversity and Evolution, Marine College, Shandong University, Weihai, China
- 3Key Laboratory of Aquatic Biodiversity and Conservation of Chinese Academy of Sciences, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China
- 4Laboratory of Eukaryotic Microbiology and Animal Biology, University of Camerino, Camerino, Italy
- 5Institute of Evolution and Marine Biodiversity, Ocean University of China, Qingdao, China
Ciliates are morphologically diverse and highly specialized unicellular eukaryotes that constitute an important component of the microbial food web. Aspidisca Ehrenberg, 1830, is a highly speciose genus that plays key ecological roles in a wide range of environments, and its species diversity has always been a hot spot in protozoan research. In this study, we investigate the living morphology, infraciliature, silverline system, and ribosomal small subunit (SSU rRNA) gene sequences of two new Aspidisca species collected from subtropical coastal waters of China using standard methods. Aspidisca spina sp. n. is characterized by having an obvious peristomial spur, two transparent posterior protrusions, seven frontoventral cirri in “polystyla-arrangement”, and six dorsal ridges. It can be distinguished from the most similar congener, A. magna Kahl, 1932, by the presence of posterior protrusions, more membranelles in the posterior part of adoral zone (AZM2), and different SSU rRNA gene sequences. A. shini sp. n. is smaller in body size, only 35–40 × 25–30 µm in vivo, with four prominent ridges on the arched dorsal side, very similar with the “well-known” species A. steini Buddenbrock, 1920, in morphological characteristics, but it can be distinguished by the arrangement of frontoventral cirri and 121 nucleotide difference in the SSU rRNA gene sequences. Phylogenetic analyses based on the SSU rRNA gene sequences revealed the systematic positions of two new taxa and supported the validity of them as distinct species.
Introduction
Euplotida is a highly specialized taxon with a huge variety of species, widely distributed in marine, freshwater, and terrestrial biotopes (Petz et al., 1995; Song & Wilbert, 2000; Song et al., 2009). In the recent classification, it contains five families: Aspidiscidae, Certesiidae, Euplotidae, Gastrocirrhidae, and Uronychiidae (Gao et al., 2016; Adl et al., 2019). The morphological characters of these ciliates tend toward simplicity, and ciliatures differentiate clearly, that is, the dorsal cilia degenerate into dorsal kineties, while the cilia on the ventral side aggregate and specialize into strong cirri (Song et al., 2009; Shao et al., 2020). This evolutionary trend has led to few morphological characters that can be reliably used as taxonomic markers, making species identification more difficult. However, the species diversity of Euplotida is much higher than expected, and about 20 new species have been added to this order in the past decade (Syberg-Olsen et al., 2016; Boscaro et al., 2019; Küppers, 2020; Lian et al., 2020; Lian et al., 2021; Serra et al., 2020; Zhang et al., 2020; Živaljić et al., 2020; Abraham et al., 2021; Valbonesi et al., 2021).
The genus Aspidisca Ehrenberg, 1830, with about 60 nominal morphospecies, is the second largest genus of Euplotida. They have highly specialized morphological characters, such as (i) oval-shaped body; (ii) rigid, generally flattened ventral surface; (iii) arched dorsal side (usually with groove and/or ridges); (iv) adoral zone separated into two parts, the anterior part highly reduced and the posterior part similar to other Euplotida taxa; (v) five strong transverse cirri, usually arranged in an oblique row; and (vi) seven (seldom eight) frontoventral cirri, regularly arranged in the anterior ventral part of the cell. With the application of silver staining in the Aspidisca taxonomy, some important morphological features were revealed, which greatly promoted the species description (Plough, 1916; Kahl, 1932; Bock, 1955; Dragesco, 1960; Dragesco, 1963; Burkovsky, 1970; Hartwig, 1973; Dragesco & Dragesco-Kernéis, 1986). About 20 Aspidisca species have been recognized using infraciliature as taxonomic character, and this genus was classified into three categories according to the number and arranged pattern of frontoventral cirri by Wu and Curds (1979), namely, “lynceus arrangement”, “polystyla arrangement”, and “satellite-like arrangement”. This separation was widely accepted by later researchers (Wu & Curds, 1979; Corliss & Snyder, 1986; Fernandez-Leborans & de Zaldumbide, 1987; Foissner et al., 1991; Song & Wilbert, 2002; Li et al., 2008; Li et al., 2010; Shen et al., 2010; Jiang et al., 2013; Lian et al., 2018). In recent years, molecular information, especially the use of the ribosomal small subunit (SSU rRNA) gene sequence, has effectively aided species classification and identification. However, sequence information of most Aspidisca species is widely missing, and for only nine species reliable gene sequences are deposited in GenBank, which not only results in a lack of support for the species identification but also hinders the progress of molecular phylogenetic analysis (Li et al., 2008; Yi et al., 2009; Li et al., 2010; Shen et al., 2010; Jiang et al., 2013; Lian et al., 2018).
In the present paper, two new Aspidisca species collected from the coastal waters of the East China Sea and were described following the basic criteria proposed by Warren et al. (2017). The living morphology, infraciliature, and silver-line system obtained from sliver staining, SSU rRNA gene sequences, and the phylogenetic relationships with congeners were provided here.
Materials and methods
Sample collection, observation, and identification
Two new species were isolated from a subtropical coastal environment near Ningbo, China (Figures 1A, B). Aspidisca spina sp. n. was collected from a sandy beach (29°46′27″N, 121°56′8″E) in Meishan Bay on 3 September 2020; the water temperature was about 32°C, and salinity was 19 PSU (Figure 1C). Aspidisca shini sp. n. was collected on 24 December 2020, from a brackish lake (29°45′3″N, 121°54′30″E); the water temperature was about 10°C, and salinity was 20 PSU (Figure 1D). Samples were taken back to the laboratory and maintained as raw culture; rice grains were added to promote the growth of bacteria as food for ciliates. A. spina sp. n. survived only a few days under laboratory conditions. A. shini sp. n. could be maintained for about 3–4 weeks.
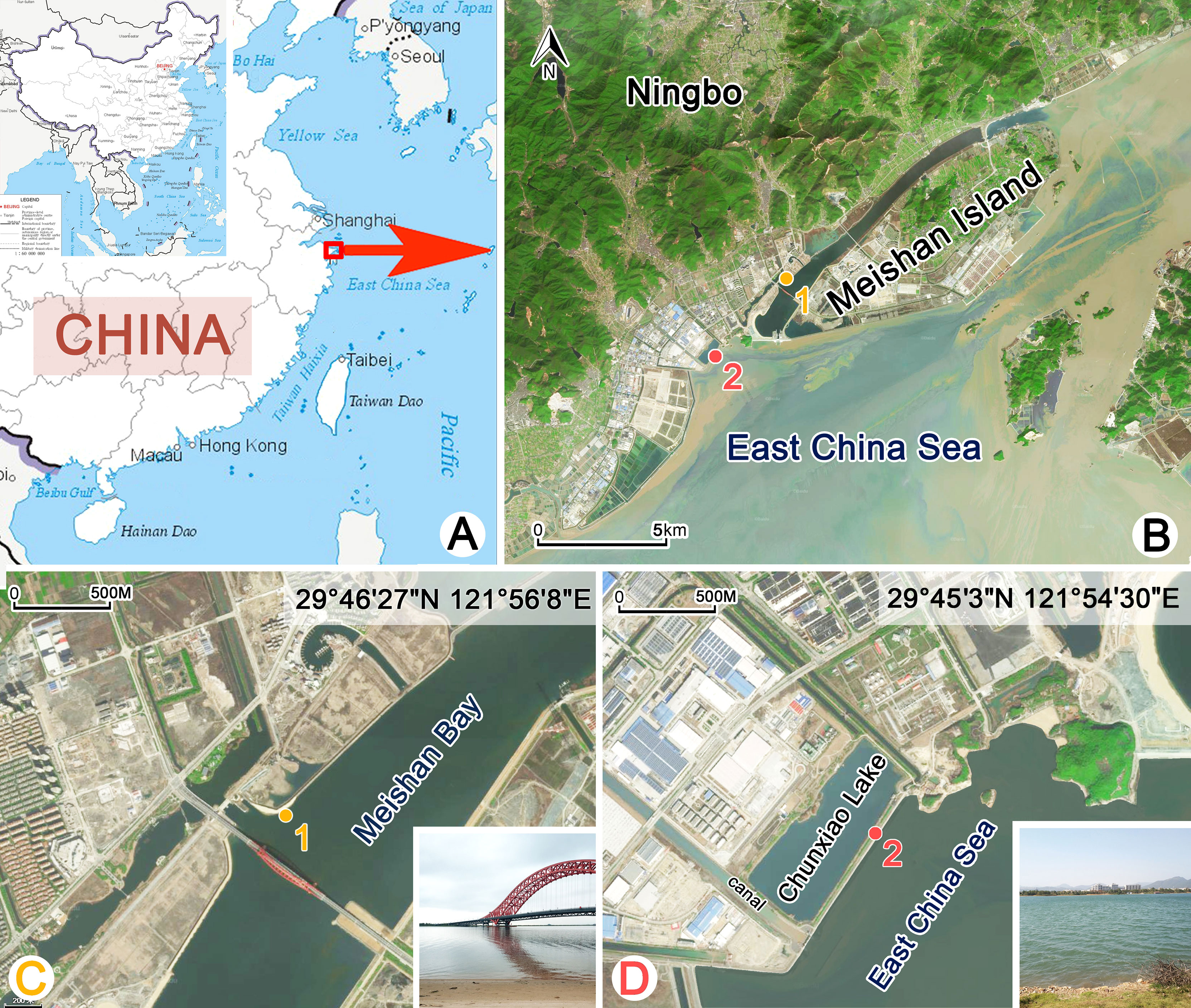
Figure 1 Locations and the habitats of sampling sites (A–D). (A, B) Map of China and the sampling sites. (C) The sandy beach near the Meishan Bay. Sampling site of Aspidisca spina sp. n. (D) Chunxiao Lake. Sampling site of Aspidisca shini sp. n.
The behaviors of two species were observed in Petri dishes using a dissecting microscope. Living cells were observed under bright field and different interference contrast (DIC) microscopy at ×400–×1,000 magnification to obtain morphological features in vivo. The ciliary pattern and nuclear apparatus were revealed using silver staining methods (Foissner, 2014; Wu et al., 2021). The protargol powder was made according to Pan et al. (2013). Counts, measurements, and drawings of stained specimens were performed at protargol powder 1,000 magnification. The terminology is mainly according to Wu and Curds (1979).
DNA extraction, gene amplification, and sequencing
The determination of the SSU rRNA gene sequences was performed on single cells for each species. The single cells were first washed with sterilized in situ water and distilled water, then transferred to a 1.5-ml microfuge tube with a minimum volume of water.
The DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) was used for single-cell DNA extractions, following the manufacturer’s instructions modified by Wang et al. (2021). Each species has a parallel sample, and the sequencing information of the two experiments was identical. The SSU rRNA gene sequence was amplified with the primers 18s-F (5′-AAC CTG GTT GAT CCT GCC AGT-3′) and 18s-R (5′-TGA TCC TTC TGC AGG TTC ACC TAC-3′) (Medlin et al., 1988). The cycling conditions were as follows: an initial denaturation at 98°C for 30 s, 35 cycles of 10 s at 98°C, 20 s at 56°C touchdown, and 100 s at 72°C, and a final extension of 5 min at 72°C (Ye et al., 2021). Q5 Hot Start High-Fidelity DNA Polymerase (New England Biolabs Co., Ltd., M0493, Beijing) was used to minimize the possibility of amplification errors. Sequencing was performed bidirectionally at Tsingke Biological Technology Company (Hangzhou, China).
Phylogenetic analyses
A phylogenetic tree was constructed by using the newly characterized SSU rRNA gene sequences and 83 related sequences in the NCBI GenBank database. Six Discocephalida species, namely, Leptoamphisiella vermis (FJ865203), Paradiscocephalus elongatus (EU684746), Pseudoamphisiella alveolata (DQ503583), Pseudoamphisiella quadrinucleata (EU518416), Prodiscocephalus borrori (DQ646880), and Discocephalus ehrenbergi (FJ196397), were selected as the outgroup taxa. The 85 SSU rRNA gene sequences were aligned using MUSCLE with default parameter settings on the GUIDANCE web server (Penn et al., 2010). BioEdit v.7.1.3.0 was used to manually trim both ends (Hall, 1999). Hypervariable sites were masked and removed using Gblocks v.0.91b with a default setting (Castresana, 2000; Talavera & Castresana, 2007). Maximum likelihood (ML) and Bayesian inference (BI) analyses were carried out online on the CIPRES Science Gateway (Edgar, 2004; Miller et al., 2010). Maximum likelihood (ML) analysis with 1,000 bootstrap iterations was constructed with RAxML-HPC2 v.8.2.11 on XSEDE (Stamatakis, 2014). Bayesian inference (BI) analysis were performed with MrBayes on XSEDE v.3.2.6 (Ronquist et al., 2012) under the GTR + I + G model selected by the program MrModeltest v.2.4 (Nylander, 2004). The chain length was 1,000,000 generations and sampled every 100 generations, whereby the first 2,500 trees were discarded as burn-in. Trees were visualized in MEGA v.7.0 (Kumar et al., 2016) and TreeView v.1.6.6 (Page, 1996). Systematic classification and terminology mainly followed Lynn (2008); Gao et al. (2016), and Lian et al. (2021).
Results
Class Spirotrichea Büetschli, 1889
Subclass Hypotrichia Stein, 1859
Order Euplotida Small & Lynn, 1985
Family Aspidiscidae Ehrenberg, 1830
Genus Aspidisca Ehrenberg, 1830
Aspidisca spina sp. n.
Diagnosis
Broadly oval body 75–80 × 50–55 µm in vivo; arched dorsal side with six ridges, a peristomial spur located on the left border, two transparent protrusions positioned posteriorly; constantly eight membranelles in the anterior portion of adoral zone (AZM1), about 20 membranelles in the posterior portion (AZM2); seven equally strong frontoventral cirri in “polystyla-arrangement”; five transverse cirri; four dorsal kineties, the left first line with about nine dikinetids, the other three lines containing about 17, 18, and 19 dikinetids, respectively; macronucleus horseshoe-shaped; brackish water habitat.
Type locality
Intertidal waters from a sandy beach (29°46′27″N, 121°56′8″E) in the Meishan Bay, Ningbo, China.
Type materials
The slide (registration number: ZXT-20200903-1-01) with the protargol-stained holotype specimen (Figures 2D, E and Figures 3G, H) has been deposited in the Laboratory of Protozoology, Ocean University of China (OUC); five protargol-stained paratype slides have been deposited in the Ningbo University (registration numbers: ZXT-20200903-1-02, 03…06).
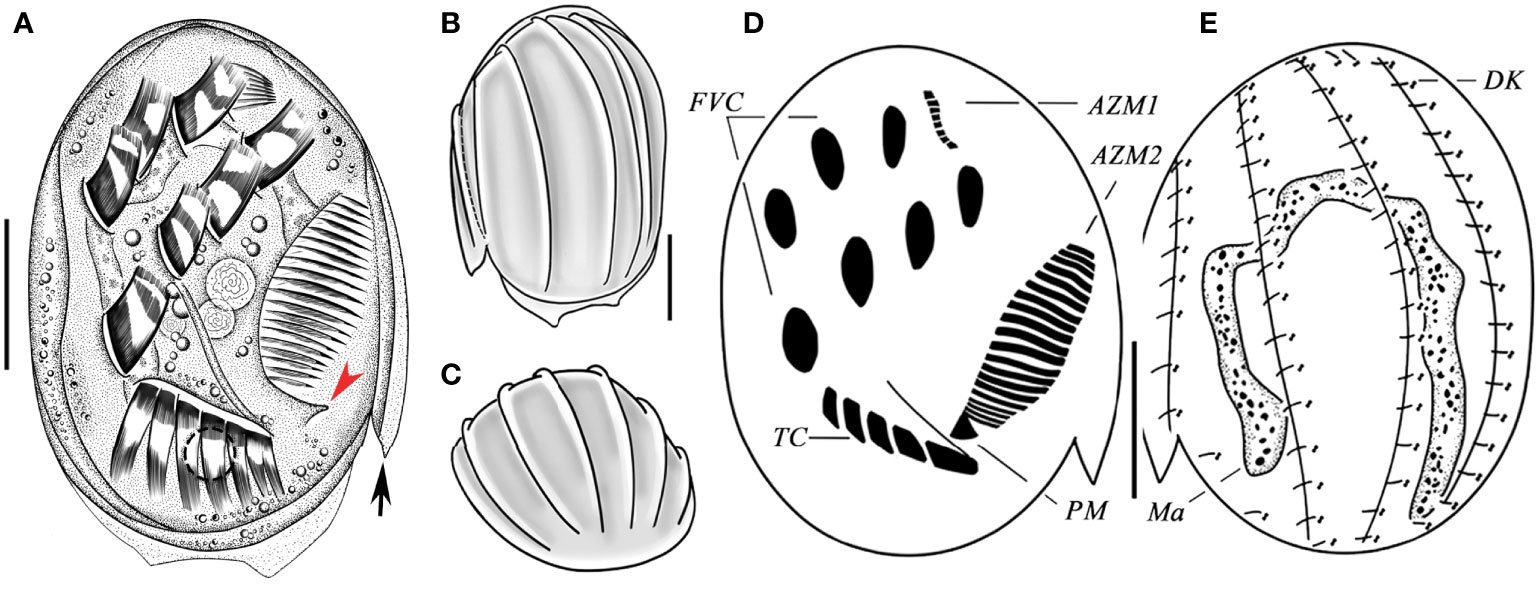
Figure 2 Aspidisca spina sp. n. from life (A–C), and after protargol staining (D, E). (A) Ventral view of a typical individual; arrow marks spur, and arrowhead marks the bulge. (B, C) Dorsal (B) and apical (C) views, showing the ridges. (D, E) Infraciliature of ventral (D) and dorsal (C) sides of the same specimen. AZM1, anterior portion of adoral zone membranelles; AZM2, posterior portion of adoral zone membranelles; DK, dorsal kineties; FVC, frontoventral cirri; Ma, macronucleus; PM, paroral membrane; TC, transverse cirri. Scale bars: 20 µm.
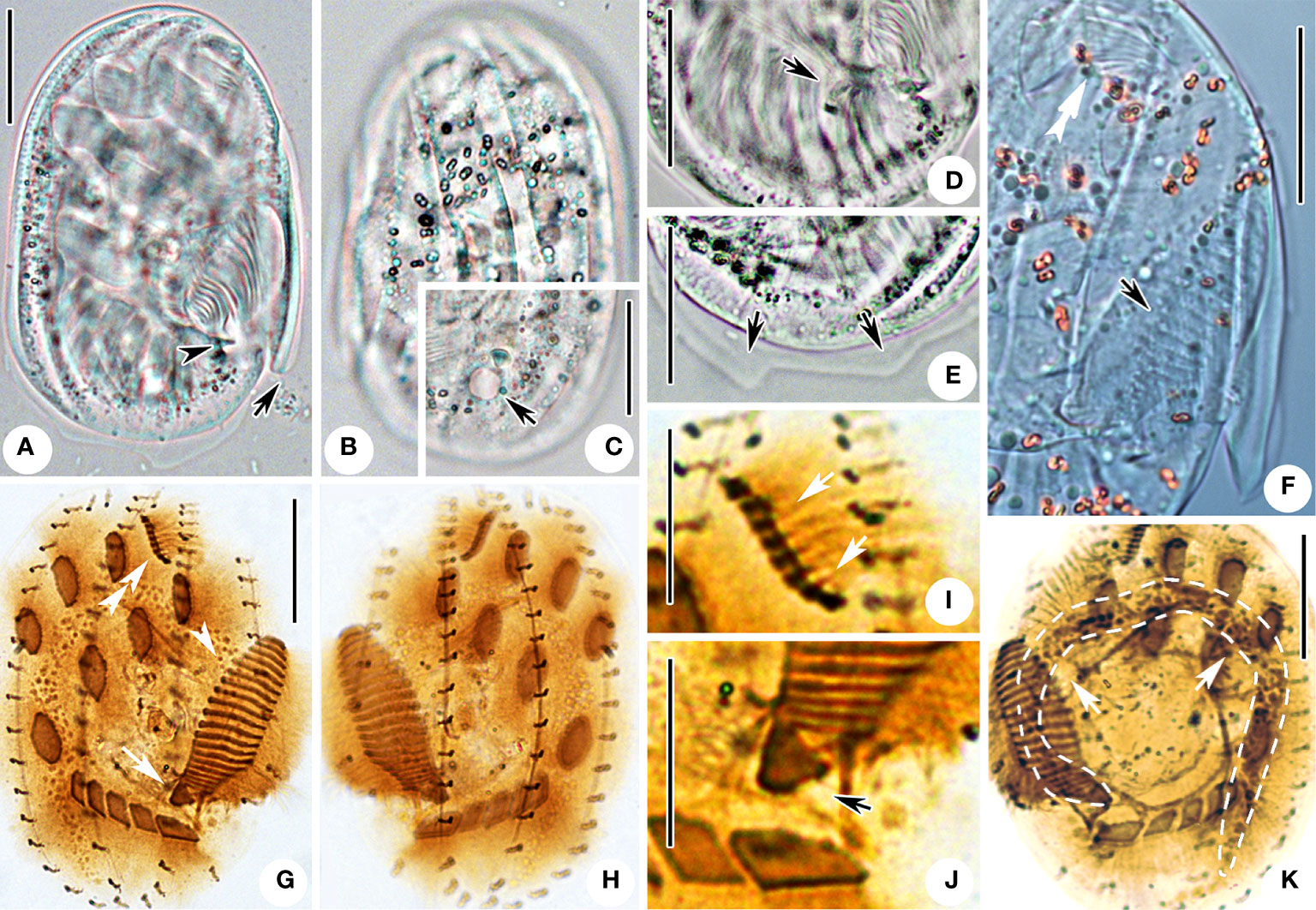
Figure 3 Microphotographs of Aspidisca spina sp. n. in life (A–F) and after protargol impregnation (G–K). (A) Ventral view of a representative individual; arrow marks the peristomial spur, and arrowhead marks the bulge. (B) Dorsal view, showing the ridges. (C) Dorsal view; arrow indicates the contractile vacuole. (D) Ventral view, arrow points to the leftmost transverse cirri. (E) Dorsal view, showing the protrusions on the posterior cell margin (arrows). (F) Ventral view; double arrowheads point to AZM1, and arrow points to AZM2. (G, H) Infraciliature of ventral and dorsal sides of the same specimen; double arrowheads point to AZM1, arrowhead points to AZM2, and arrow points to PM. (I) Ventral view, arrows showing the AZM1. (J) Ventral view of the posterior part of AZM2; arrow marks the paroral membrane. (K) Dorsal view, arrows showing the macronucleus. Scale bars: 20 µm (A–H, K), 10 µm (I, J).
Etymology
The Latin adjective “spina” refers to the prominent spur near peristomial area.
ZooBank registration
urn:lsid:zoobank.org:act:A4C7A755-8A58-4DB3-8C75-0D95A41A3884
Morphological description
Cell size 75–80 × 50–55 µm in vivo, and about 52–73 × 39–61 µm after protargol impregnation, body shape broadly oval, right edge straight and smooth, left border with one beak-like peristomial spur located near the posterior part of AZM2 (Figures 2A, arrow, 3A, F), the posterior cell hyaline and margin with two transparent protrusions (Figures 2A, 3E, arrow). Dorsoventrally flattened about 3:1. Dorsal surface usually with six longitudinal ridges, central four more prominent (Figures 2B, C, 3B). Buccal field ellipsoidal with right border clearly indented formed a cambered cavity, an obvious bulge near posterior end of oral opening (Figures 2A, arrowhead, 3A, arrowhead). Cytoplasm colorless, transparent around cell periphery, slightly opaque in central part due to many inclusions (about 2 µm in diameter). Contractile vacuole about 6 µm in diameter when fully extended, located behind the leftmost transverse cirrus (Figure 3C, arrow). Macronucleus horseshoe-shaped with rounded ends (Figure 3K). Micronucleus not observed in vivo nor in stained specimens.
Ciliary pattern as shown in Figures 2D, E, 3G, H. Invariably seven frontoventral cirri (FVC) in “polystyla-arrangement” rather stiff and strong, about 15 µm long. Transverse cirri (TC) strong, tightly arranged in oblique row, about 13–17 µm long, the leftmost one largest and composed of three cirri in vivo (Figures 2D, 3D, G, J). The anterior portion of adoral zone (AZM1) always composed of eight membranelles, located in shallow concave (Figures 3F, I; Table 1), the posterior part (AZM2) including 19–21 membranelles, the cilia about 10–15 µm long (Figures 3A, F; Table 1). Paroral membrane small, arranged in area between the adoral zone and the leftmost transverse cirri (Figure 2D). Four dorsal kineties (DK), each with a relatively constant number of basal body pairs, from left to right consisting of about 9, 17, 18, 19 bristles respectively. Each kinety generally extends from one end to the other end of the cell (Figures 2E, 3H; Table 1).
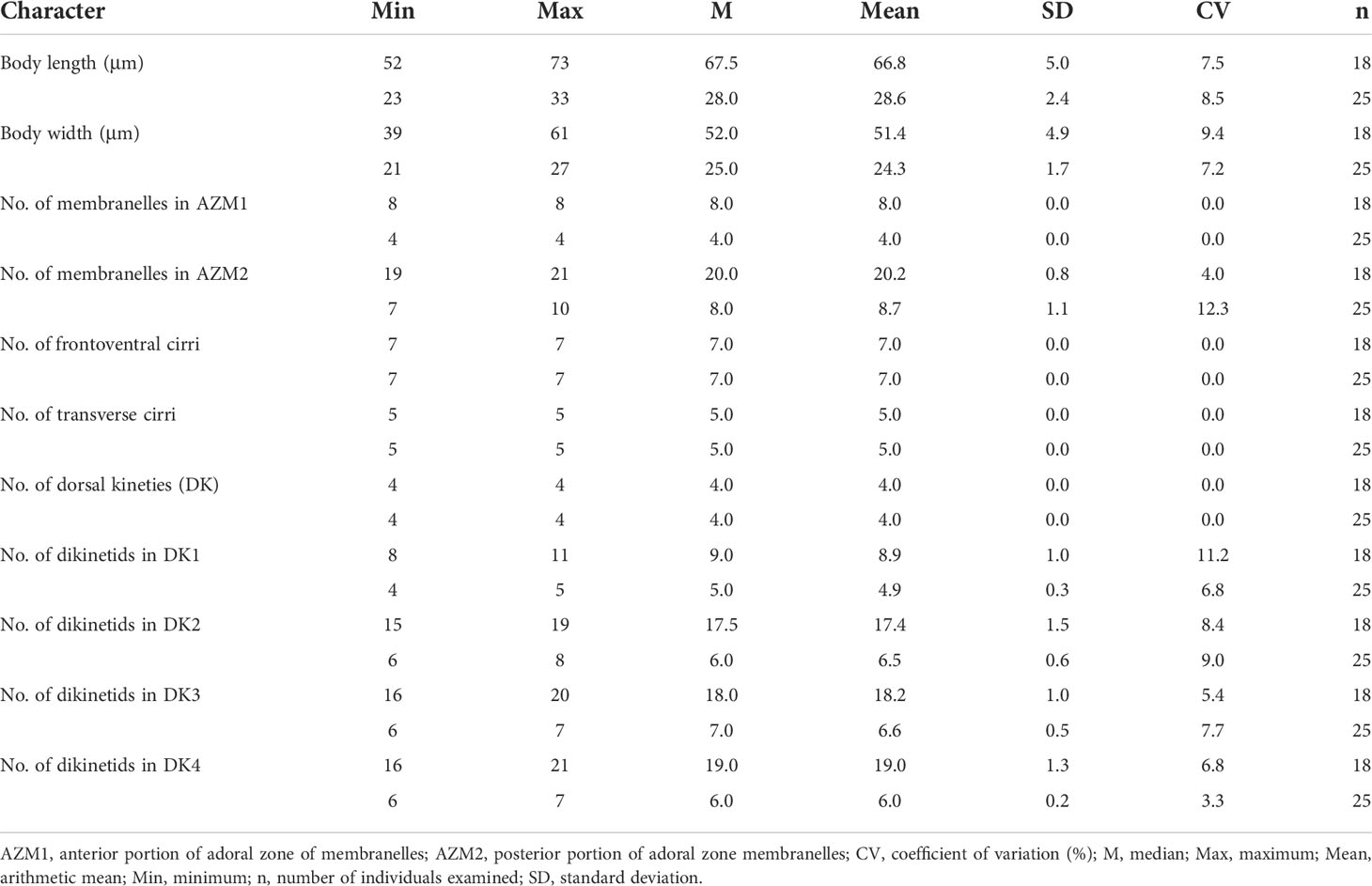
Table 1 Morphometric data of Aspidisca spina sp. n. (upper line) and A. shini sp. n. (lower line) based on protargol-stained specimens.
Aspidisca shini sp. n.
Diagnosis
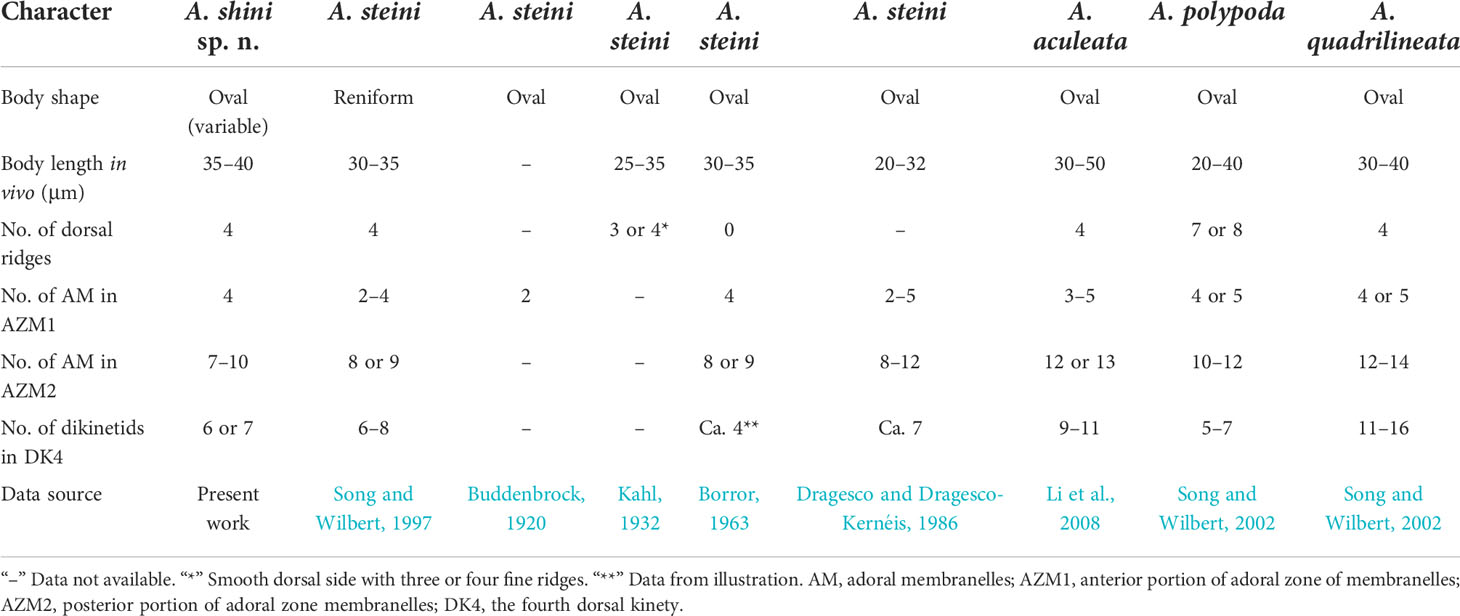
Broadly oval body 35–40 × 25–30 µm in vivo, slightly arched dorsal side with four dorsal ridges; four membranelles in the anterior portion of adoral zone (AZM1), 7–10 in the posterior portion (AZM2); seven equally strong frontoventral cirri arranged in “polystyla-arrangement”, five transverse cirri; four dorsal kineties, from left to right every kinety with on average 5, 7, 7, 6 dikinetids respectively; macronucleus typically horseshoe-shaped; brackish water habitat.
Type locality
A brackish artificial lake, named Chunxiao Lake (29°45′3″N, 121°54′30″E), near the coast of the East China Sea, in Ningbo, China.
Type materials
The slide (registration number: ZBH-20201224-1-03) with the protargol-stained holotype specimen (Figures 4D, E, 5I, J) has been deposited in the Laboratory of Protozoology, Ocean University of China (OUC); seven paratype slides have been deposited in Ningbo University, including five protargol-stained slides (registration numbers: ZBH-20201224-1-01, 02, 04…06) and two silver nitrate-stained slides (registration numbers: ZBH-20201224-1-07, 08).
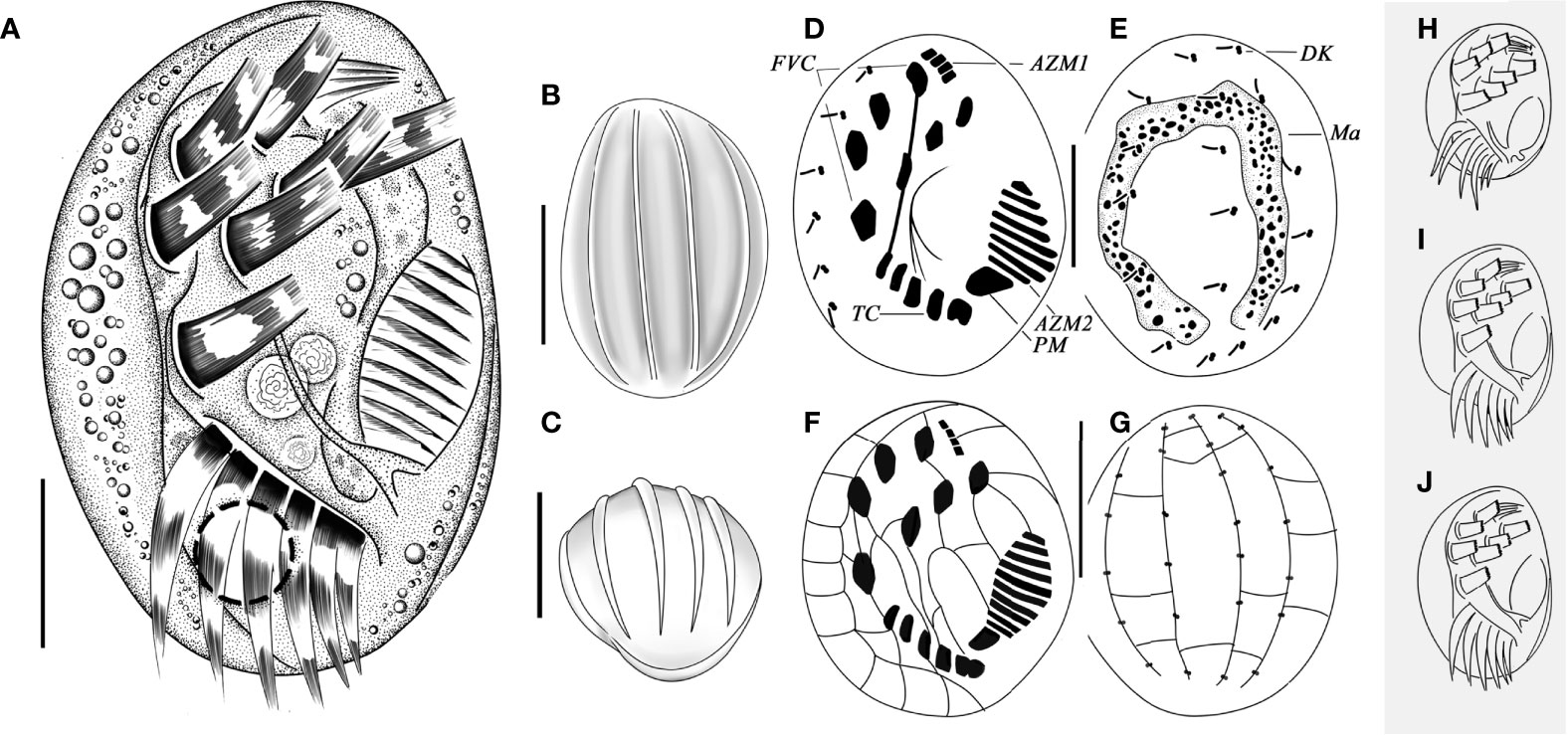
Figure 4 Living morphology (A–C, H–J), infraciliature (D, E), and silverline system (F, G) of Aspidisca shini sp. n. (A) Ventral views of a normal individual. (B, C) Dorsal (B) and apical (C) views, showing the ridges. (D, E) Infraciliature of ventral (D) and dorsal (E) sides of the specimens. (F, G) Silverline system on dorsal (F) and ventral (G) sides. (H–J) Ventral view of cells with different shapes. AZM1, anterior portion of adoral zone of membranelles; AZM2, posterior portion of adoral zone membranelles; DK, dorsal kineties; FVC, frontoventral cirri; Ma, macronucleus; PM, paroral membrane; TC, transverse cirri. Scale bars: 20 µm (B, C, F, G), 10 µm (A, D, E).
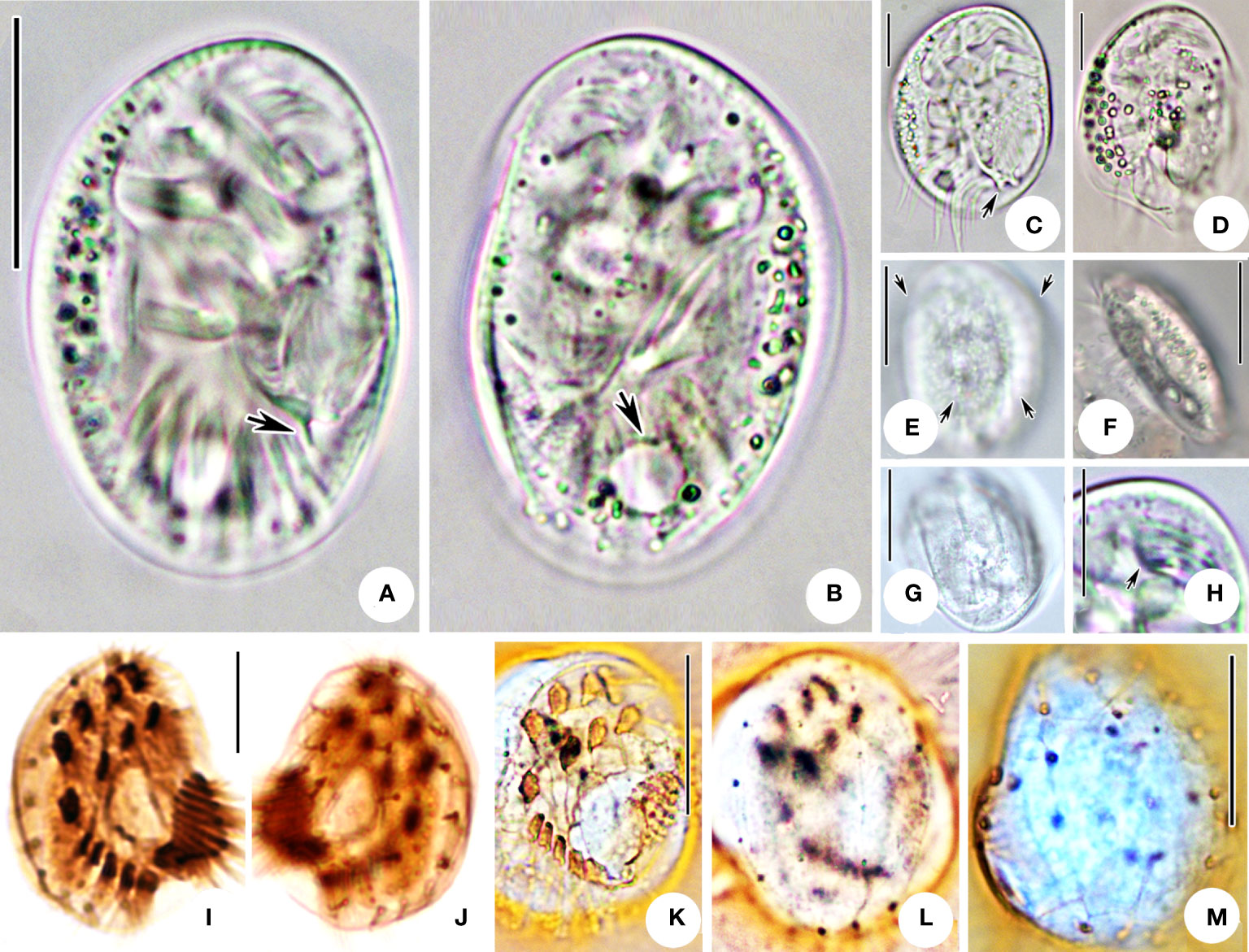
Figure 5 Microphotographs of Aspidisca shini sp. n. in life (A–H), after protargol impregnation (I, J) and silver nitrate impregnation (K–M). (A) Ventral view of a representative individual; arrow indicates the protrusion. (B) Dorsal view, arrow indicates the contractile vacuole. (C, D) Cells with different shapes; arrow indicates the protrusion. (E) Dorsal view; arrows indicate the ridges. (F, G) Lateral (F) and apical (G) views of two individuals. (H) Ventral view of the anterior part of the cell, arrow showing the AZM1. (I, J) Infraciliature of ventral (I) and dorsal (J) sides of the specimens. (K–M) Silver nitrate impregnation of ventral (K) and dorsal (L, M) sides. Scale bars: 20 µm (A, B, E–G, K–M), 10 µm (C, D, H–J).
Etymology
We dedicate this new species to Professor Mann Kyoon Shin, in recognition of his great contributions to the ciliatology studies.
ZooBank registration
urn:lsid:zoobank.org:act:2804058C-588E-4096-82B8-EC089465A441
Morphological description
Body about 35–40 × 25–30 µm in vivo, and about 22–33 × 21–27 µm after protargol impregnation, oval-shaped, right side slightly arched near the oral area, left side curved in arc (Figures 5A, B). Body shape slightly variable after cultured several days (Figures 4H–J, 5C, D). Dorsoventrally flattened about 5:2 (Figures 5E, F), with the ventral surface generally flat and a slightly arched dorsal surface invariably with four low ridges (Figures 4B, C, 5E, arrow, G). Buccal field ellipsoidal, a triangle-shaped protrusion near the posterior end of the adoral area (Figures 5A, arrow, C, arrow). Cytoplasm colorless and hyaline. Right part of cell usually filled with a large number of granules and oil droplets (Figures 5A–D). Contractile vacuole about 6 µm in diameter, located behind transverse cirri. Horseshoe-shaped macronucleus. Micronucleus not observed in vivo nor in stained specimens.
Ciliary pattern as shown in Figures 4D, E, 5I, J. There are seven equally strong frontoventral cirri (FVC) in “polystyla-arrangement”, each cilium about 9 µm long, five transverse cirri tightly arranged in an oblique row, the leftmost cirrus always split into two parts slightly (Figures 5A,C). Cilia of transverse cirri about 10–15 µm long. The anterior portion of adoral zone (AZM1) composed four membranelles located in deep depression (Figure 5H; Table 1), the posterior part of adoral zone (AZM2) composed of 7–10 membranelles (Figures 4A, D; Table 1), cilia about 8–11 µm long. Paroral membrane (PM) conspicuous and big, arranged in area between the adoral zone and the leftmost transverse cirri (Figure 5I). Invariably four dorsal kineties with sparsely arranged basal body pairs, from left to right consisting of about five, seven, seven, and six dikinetids, respectively. Each kinety aligned with a dorsal ridge and extending entire length of the cell (Figures 5J, L; Table 1). Silverline system on ventral side following typical genus pattern (Figures 4F, 5K), dorsal argyrome simple, consisting of four longitudinal primary meridians which connected to dikinetids (Figures 4G, 5L, M).
Molecular data and phylogenetic analyses
The newly obtained SSU rRNA gene sequences were submitted to the GenBank database with the length (bp), G + C content, and accession numbers as follows: Aspidisca spina sp. n., 1,500 bp, 48.67%, OM952922; and A. shini sp. n., 1,514 bp, 45.57%, OM952923.
The topologies of the maximum likelihood (ML) and Bayesian inference (BI) trees were almost the same; therefore, only the ML tree is shown here with nodal support values from both algorithms (Figure 6). In the phylogenetic tree, the family Aspidiscidae is monophyletic as all members grouped into one well-supported clade (ML 96%, BI 1.00). It forms a robust sister relationship to the cluster of Euplotidae and Certesiidae (ML 100%, BI 1.00). Within the Aspidiscidae clade, the new species A. spina sp. n. is closely related to A. magna (EU880598) with moderate support (ML 79%, BI 0.98). A. shini sp. n. groups with A. aculeata (EF123704) with full support (ML 100%, BI 1.00), followed by A. orthopogon (EU430745).
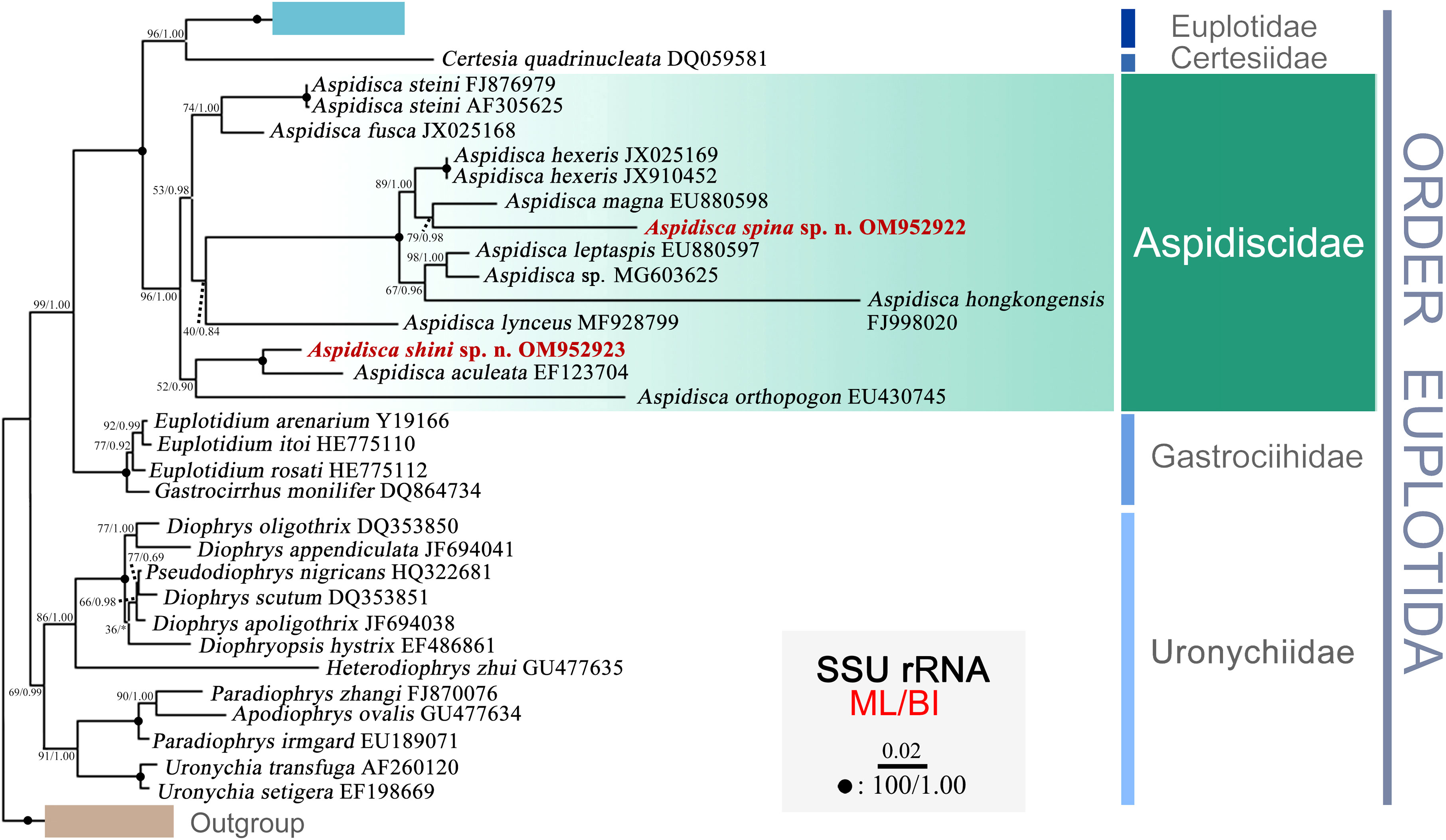
Figure 6 Phylogenetic tree inferred from SSU rRNA gene sequences, using maximum likelihood (ML) and Bayesian inference (BI) methods. Sequences of the new described species are highlighted in bold and red. Bootstrap support for ML tree and posterior probability for BI tree are given near nodes. Fully supported (100/1.00) branches are marked with solid circles. The scale bar represents two substitutions per 100 nucleotide positions.
Discussion
Comments on Aspidisca spina sp. n.
The distinctive features of Aspidisca spina sp. n. are the beak-like peristomial spur and two transparent protrusions on the posterior end of the cell. In terms of the conspicuous peristomial spur and the “polystyle-arrangement” of frontoventral cirri, five species should be compared with A. spina sp. n., that is, A. magna Kahl, 1932, A. sedigita Quennerstedt, 1867, A. hexeris Quennerstedt, 1869, A. fusca Kahl, 1928, and A. dentata Kahl, 1928 (Table 2).
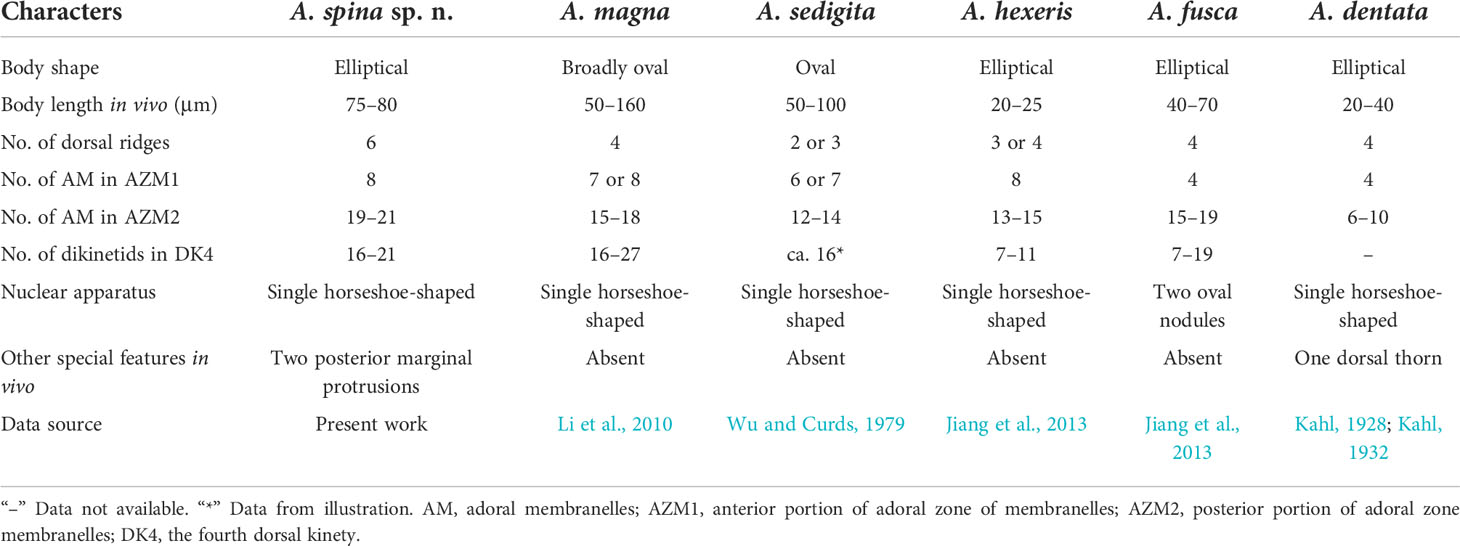
Table 2 Comparison of Aspidisca spina sp. n. with some congeners having peristomial spur and seven frontoventral cirri in “polystyla-arrangement”.
Aspidisca magna was firstly collected from Kiel by Kahl (1932). Based on the Qingdao population, Li et al. (2010) redescribed this species in detail using modern research methods. It is very similar to A. spina sp. n. in the body shape, arrangement of transverse cirri, number of membranelles in AZM1, and dikinetids in the dorsal kineties. However, these two species can be clearly separated by the protrusion structure (absent in A. magna vs. two posterior marginal protrusions in A. spina sp. n.), the number of dorsal ridges (4 in A. magna vs. 6 in A. spina sp. n.), and membranelles in AZM2 (15–18 in A. magna vs. 19–21 in A. spina sp. n.) (Figures 7A–C; Table 2). In addition, the SSU rRNA gene sequences of the two species differ from each other by 159 nucleotides (Li et al., 2010).
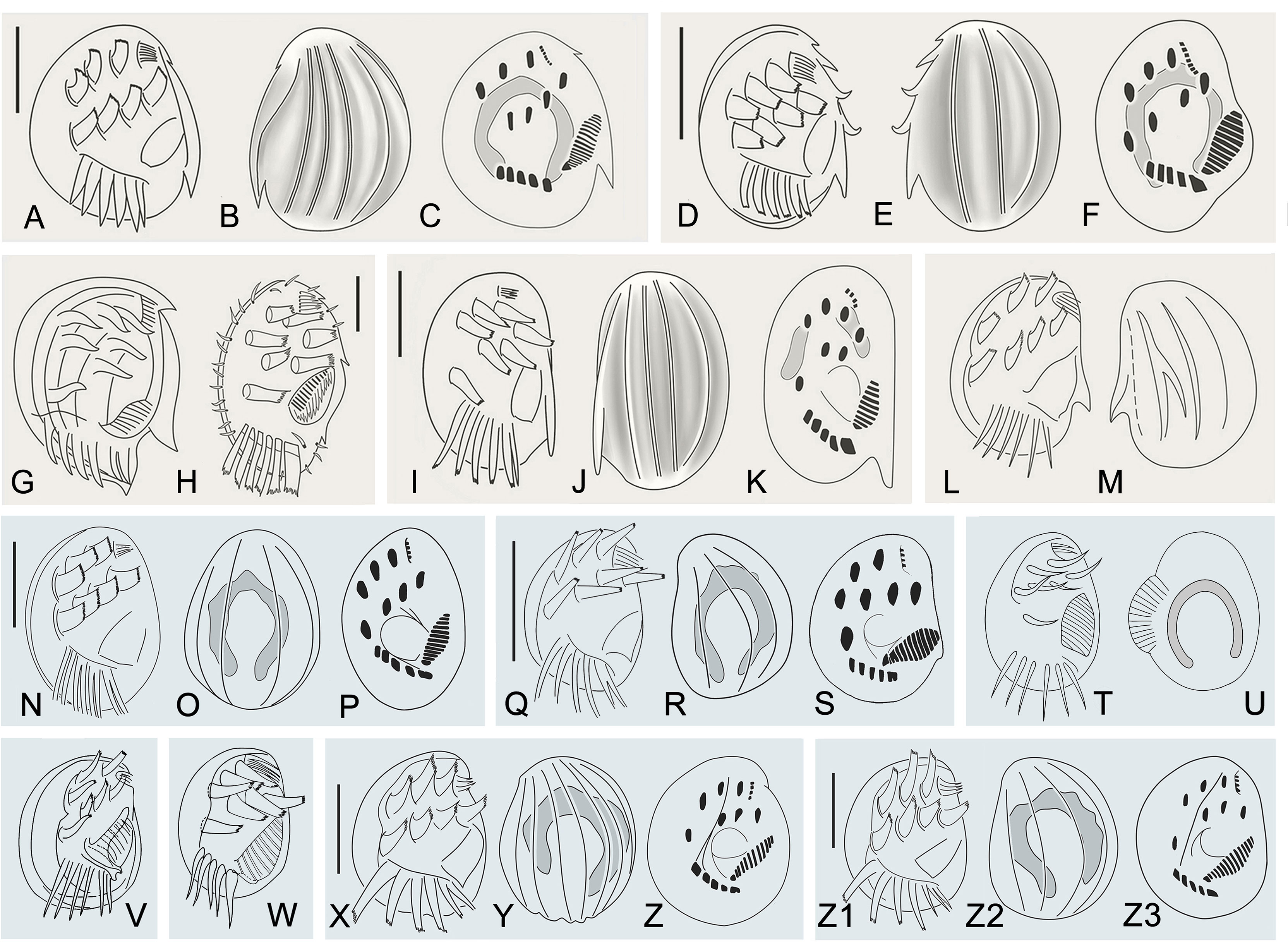
Figure 7 Morphological characters of Aspidisca species are similar to Aspidisca spina sp. n. (A–M) and A. shini sp. n. (N–Z3). (A–C) Aspidisca magna (redrawn after Li et al., 2010). (D–F) Aspidisca hexeris (redraw after Jiang et al., 2013). (G, H) Aspidisca sedigita (M, redrawn after Quennerstedt, 1867; (N), redrawn after Dragesco, 1963). (I–K) Aspidisca fusca (redrawn after Jiang et al., 2013). (L, M) Aspidisca dentata (redrawn after Kahl, 1932). (N–P) Aspidisca aculeata (redrawn after Li et al., 2008). (Q–W) Aspidisca steini (Q–S), redrawn after Song & Wilbert, 1997; (T, U), redrawn after Buddenbrock, 1920; (V), redrawn after Kahl, 1932; (W), redrawn after Borror, 1963). (X–Z) Aspidisca polypoda (redrawn after Song & Wilbert, 2002). (Z1–3) Aspidisca quadrilineata (redrawn after Song &Wilbert, 2002). Scale bars: 40 µm (A–C), 20 μm (D–F, H–K, N–S, X–Z3).
Aspidisca sedigita was initially found by Quennerstedt (1867) in Visby, Gotland Island; Wu and Curds (1979) made a short review of this species. A. sedigita and A. spina sp. n. share the same nuclear apparatus (single horseshoe-shaped macronucleus) and the presence of posterior protrusion (population of A. sedigita in Quennerstedt’s original report also with posterior protrusions, but not obvious in other populations). A. sedigita differs from the new species in the number of dorsal ridges (2 or 3 vs. 6) and membranelles in AZM1 (6 or 7 vs. 8) and in AZM2 (12–14 vs. 19–21). (Figures 7G, H; Table 2) (Dragesco, 1963; Agamaliev, 1967; Wu & Curds, 1979).
Aspidisca hexeris was also collected from Visby, Gotland Island, by Quennerstedt (1869) and was regarded as synonym of A. sedigita in the revision of Wu and Curds (1979). Jiang et al. (2013) clarified the taxonomy position of this species based on the comprehensive morphological and molecular information. It resembles A. spina sp. n. in the body shape, eight membranelles in AZM1, and the horseshoe-shaped macronucleus. However, A. hexeris can be separated from the A. spina sp. n. by the smaller body length (20–25 µm vs. 75–80 µm), smooth cell ends (vs. two posterior marginal protrusions in A. spina sp. n.), fewer dorsal ridges (3 or 4 vs. 6), fewer membranelles in AZM2 (13–15 vs. 19–21), and fewer dikinetids in the fourth dorsal kinety (7–11 vs. 16–21) (Figures 7D–F; Table 2). Furthermore, A. hexeris can be distinguished from A. spina sp. n. by the 136-nucleotide substitutions in the SSU rRNA gene sequences (Jiang et al., 2013).
Aspidisca fusca was originally collected from Bad Oldesloe, Germany, by Kahl (1928), and the infraciliature was firstly provided by Jiang et al. based on a Chinese population (2013). Although A. fusca also has a prominent peristomial spur, it differs from A. spina sp. n. for the absence of posterior protrusions, the fewer membranelles in AZM1 (4 vs. 8) and AZM2 (15–19 vs. 19–21), and the morphology of the macronucleus (two sausage nodules vs. single horseshoe-shaped). (Figures 7I–K, Table 2). Additionally, A. fusca is genetically distinct from A. spina sp. n. by 197-nucleotide substitutions in the SSU rRNA gene sequences (Jiang et al., 2013).
Aspidisca dentata was firstly discovered by Kahl (1928) but has not been studied in more detail since then. It is similar with A. spina sp. n. in the elliptical body shape and single horseshoe-shaped macronucleus. However, A. dentata has fewer dorsal ridges (4 vs. 6 in A. spina sp. n.) and fewer membranelles in AZM1 (4 vs. 8 in A. spina sp. n.) and AZM2 (6–10 vs. 19–21 in A. spina sp. n.). Besides, A. dentata has a prominent dorsal thorn (vs. absence in A. spina sp. n.), but A. spina sp. n. has two posterior protrusions (absence in A. dentata) (Figures 7L, M; Table 2) (Kahl, 1928; Kahl, 1932).
Comment on Aspidisca shini sp. n.
Aspidisca shini sp. n. is a brackish water species with a small size. It is mainly characterized by the oval-shaped outline, the “polystyla-arrangement” frontoventral cirri, and the horseshoe-shaped macronucleus. With reference to its morphological characteristics and ciliature, it can be compared with A. steini Buddenbrock, 1920, A. aculeata (Ehrenberg, 1838) Kahl, 1932, A. polypoda Kahl, 1932, and A. quadrilineata Kahl, 1932.
Aspidisca steini is a well-known species first found in a marine aquarium in Germany (Buddenbrock, 1920). Although the original description was very brief and incomplete, the diagrams provided the accurate arrangement of ventral cirri (Figures 7T, U). About 10 years later, Kahl (1932) identified an Aspidisca collected from Germany (Kiel) as this species and presented some morphological information; however, the arrangement of frontoventral cirri was slightly different from the original description, that is, two lines arranged in two parallel rows (Figure 7V) (Kahl, 1932). Borror (1963) provided the ciliary pattern of this species for the first time based on a population collected from USA (Figure 7W). The arrangement of frontoventral cirri was the same as Kahl’s population (Borror, 1963). Subsequently, several populations from Africa were investigated by Dragesco and Dragesco-Kernéis (1986) and more taxonomic data were provided. Unfortunately, those morphological data were unstable, including the arrangement of the frontoventral cirri, the number of membranes in AZM1, and the number of dorsal kineties, and even the number of transverse cirri was not stable. Thus, we suggest that Dragesco and Dragesco-Kernéis’ isolation is likely a complex Aspidisca species (Dragesco & Dragesco-Kernéis, 1986). The detailed morphological description of this species was provided by Song and Wilbert (1997) based on a population collected from China, and the molecular information was also supplied by the same laboratory several years later (Chen & Song, 2002). The arrangement of frontoventral cirri of a China population was very similar with the original population (Figures 7Q–S). Till now, the China population should be the most well-described population of A. steini (Song & Wilbert, 1997; Chen & Song, 2002). Therefore, we mainly compare the new species with the China population of A. steini in this work. It is very similar with A. shini sp. n. in the body size, number of dorsal ridges, number of membranelles in AZM1 and AZM2, and number of dikinetids in the dorsal kinety. However, the former can be clearly distinguished from the latter by the arrangement of the frontoventral cirri (the second line arranged in curved arc, name cirrus III/2 not in the same line as the others vs. approximately arranged in two parallel rows) (Figures 7Q–S; Table 3). Furthermore, A. steini is genetically distinct from A. shini sp. n. by 121-nucleotide substitutions in the SSU rRNA gene sequences. Based on the arrangement of frontoventral cirri, we conclude that only the China population of A. steini is identical to the original description; the populations from USA and Germany (Kiel) of A. steini should be synonymized with A. shini n. sp.
Aspidisca aculeata was discovered by Ehrenberg (1838) in Kiel and then has been frequently reported in the later literature; the latest comprehensive description was provided by Li et al. (2008). It is similar with A. shini sp. n. in cell size and number of dorsal ridges and membranelles in AZM1. However, the former can be separated from the latter by the number of membranelles in AZM2 (12 or 13 vs. 7–10) and dikinetids in the fourth dorsal kinety (9–11 vs. 6 or 7) (Figures 7N–P; Table 3). In addition, the two species differs from each other by 95-nucleotide substitutions in the SSU rRNA gene sequences (Li et al., 2008).
Aspidisca polypoda, originally found by Dujardin (1841), was redescribed by Song and Wilbert (2002) and Choi et al. (2020). It is very similar to A. shini sp. n. in body size, number of membranelles in AZM1, and number of dikinetids in the fourth dorsal kinety. However, these two species have a different number of dorsal ridges (7 or 8 in A. polypoda vs. 4 in A. shini sp. n.) and membranelles in AZM2 (10–12 in A. polypoda vs. 7–10 in A. shini sp. n.). Moreover, the frontoventral cirri of the new species are more stout and powerful than cirri of A. polypoda (Figures 7X–Z, Table 3) (Song & Wilbert, 2002; Choi et al., 2020).
Aspidisca quadrilineata was firstly described by Kahl (1932); Song and Wilbert (2002) improved the former work by providing the detailed description and the silverline system. A. quadrilineata shares many morphological features with A. shini sp. n., such as cell size, number of dorsal ridges, and number of membranelles in AZM1. In addition to the thin and delicate frontoventral cirri, A. quadrilineata differs from A. shini sp. n. by the higher number of membranelles in AZM2 (12–14 vs. 7–10) and dikinetids in the fourth dorsal kinety (11–16 vs. 6 or 7) (Figures 7Z1–Z3, Table 3) (Song & Wilbert, 2002).
Phylogenetic analyses based on SSU rRNA gene sequences
As shown in Figure 6, the Order Euplotida is a well-supported clade that contains five families, namely, Euplotidae, Certesiidae, Aspidiscidae, Gastrociihidae, and Uronychiidae, which has been recognized in previous studies (Yi et al., 2009; Shen et al., 2010; Huang et al., 2011; Jiang et al., 2013; Lian et al., 2018). The internal relationships in the monophyletic family Aspidiscidae are far from robust, as indicated by the low support values. The previously proposed criteria to group members of the genus Aspidisca according to their freshwater or marine habitat, as proposed by Kahl (1932), or on the basis of number and pattern of frontoventral cirri, as proposed by Wu and Curds (1979), are not supported by molecular phylogeny. Indeed, the two new species here described were both collected from the coastal waters of the East China Sea and share the same frontoventral pattern of “polystyla-arrangement” with other species as well; however, they do not cluster together within the same clade of the phylogenetic tree. This finding suggests that the morphology-based classification of Aspidisca can be problematic, particularly when species have few morphological characters and even the use of silver staining or transmission electron microscopy does not add useful information. Therefore, SSU rRNA gene sequence phylogeny may represent an important tool that, together with morphology, could help to construct a more valuable classification system. Since only 12 SSU rRNA gene sequences of the approximately 60 Aspidisca morphospecies are available, the correlation between detailed morphological data and gene sequences for a greater number of taxa is thus required to determine more stable phylogenetic relationships within the family Aspidiscidae.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
BZ and XZ were involved in the sampling, cultivation, and staining of the ciliates. Next-generation sequencing data were generated by BZ and XZ and analyzed by TY. The phylogenetic analysis was performed by BZ, JH, and YJ. BZ, TY, and AV wrote the manuscript, and XC supervised the studies. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Science Foundation of China (Project number: 31970398) and the Youth Innovation Promotion Association of the Chinese Academy of Sciences (No. 2019333).
Acknowledgments
The authors are very thankful to Weibo Song for improving the quality of figures.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abraham J. S., Somasundaram S., Maurya S., Gupta R., Makhija S., Toteja R. (2021). Characterization of Euplotes lynni nov. spec., E. indica nov. spec. and description of E. aediculatus and E. woodruffi (Ciliophora, euplotidae) using an integrative approach. Eur. J. Protistol. 79, 125779. doi: 10.1016/j.ejop.2021.125779
Adl S. M., Bass D., Lane C. E., Lukeš J., Schoch C. L., Smirnov A., et al. (2019). Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 66, 4–119. doi: 10.1111/jeu.12691
Agamaliev F. G. (1967). Faune des ciliés mésopsammiques de lacôte ouest de la mer caspienne. Cah. Biol. Mar. 8, 359–402.
Bock K. (1955). Condylostoma vastum nov. spec., und Aspidisca pertinens nov. spec., zwei sandbewohnende ciliaten aus dem küstengebiet der kieler bucht. Zool. Anz. 154, 302–304.
Borror A. C. (1963). Morphology and ecology of the benthic ciliated protozoa of alligator harbor, Florida. Arch. Protistenk. 106, 465–534.
Boscaro V., Syberg-Olsen M. J., Irwin N. A., del Campo J., Keeling P. J. (2019). What can environmental sequences tell us about the distribution of low-rank taxa? the case of Euplotes (Ciliophora, spirotrichea), including a description of euplotes enigma sp. nov. J. Eukaryot. Microbiol. 66, 281–293. doi: 10.1111/jeu.12669
Buddenbrock W. (1920). Beobachtungen über einige neue oder wenig bekannte marine infusorien. Arch. Protistenk. 41, 341–364.
Burkovsky I. V. (1970). The ciliates of the mesopsammon of the kandalaksha gulg (White sea). I. Acta Protozool. 7, 475–489.
Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Chen Z., Song W. (2002). Phylogenetic positions of Aspidisca steini and Euplotes vannus within the order euplotida (Hypotrichia: Ciliophora) inferred from complete small subunit ribosomal RNA gene sequences. Acta Protozool. 41, 1–9. doi: 10.1016/S0378-1135(01)00446-1
Choi J. M., Jung J. H., Kang J. H., Kim Y. O. (2020). New record of four ciliates (Protozoa, ciliophora) collected from rocky intertidal pools of south Korea. J. Species. Res. 9, 455–461. doi: 10.12651/JSR.2020.9.4.455
Corliss J. O., Snyder R. A. (1986). A preliminary description of several new ciliates from the Antarctica, including Cohnilembus grassei n. sp. Protistologica 22, 39–46.
Dragesco J. (1960). Ciliés mésopsammiques littoraux, systématique, morphologie, écologie. Trav. Stn. Biol. Roscoff. 12, 1–356.
Dragesco J. (1963). Compléments a la connaissance des ciliés mésopsammiques de roscoff. Cah. Biol. Mar. 4, 251–275.
Dragesco J., Dragesco-Kernéis A. (1986). Ciliés libres de l’Afrique intertropicale. introduction à la connaissance et à l’étude des ciliés. Faune Trop. 26, 1–559.
Dujardin F. (1841). Histoire Naturelle des Zoophytes Infusoires. Libraire Encyclopédique de Roret, Paris.
Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.2460/ajvr.69.1.82
Ehrenberg C. G. (1838). Die infusionsthierchen als vollkommene organismen (Voss, Leipzig: L. Germany).
Fernandez-Leborans G., de Zaldumbide M. C. (1987). A new species of the genus Aspidisca (Ciliophora, hypotrichida). J. Nat. Hist. 21, 1293–1301. doi: 10.1080/00222938700770791
Foissner W. (2014). An update of ‘basic light and scanning electron microscopic methods for taxonomic studies of ciliated protozoa’. Int. J. Syst. Evol. Microbiol. 64:271–292. doi: 10.1099/ijs.0.057893-0
Foissner W., Blatterer H., Berger H., Kohmann F. (1991). Taxonomische und ökologische revision der ciliaten des saprobiensystems band 1: Cyrtophorida, oligotrichida, hypotrichia, colpodea (Munich: Informationsberichte des Bayer. Landesamtes. Wasserwirtschaft), 478.
Gao F., Warren A., Zhang Q., Gong J., Miao M., Sun P., et al. (2016). The all-data-based evolutionary hypothesis of ciliated protists with a revised classification of the phylum ciliophora (Eukaryota, alveolata). Sci. Rep. 6, 24874. doi: 10.1038/srep24874
Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. doi: 10.1021/bk-1999-0734.ch008
Hartwig E. (1973). Die ciliaten des gezeiten-sandstrandes der nordseeinsel sylt II: Ökologie. Mikrofauna Meeresboden. 18, 385–453.
Huang J., Yi Z., Miao M., Al-Rasheid K. A. S., Song W. (2011). Phylogenetic relationships within the genus Aspidisca (Protozoa, ciliophora, euplotida) revealed by ITS1-5.8S-ITS2 region sequences. Chin. J. Oceanol. Limnol. 29, 277–283. doi: 10.1007/s00343-011-0127-2
Jiang J., Huang J., Li J., Al-Rasheid K. A. S., Al-Farraj S. A., Lin X., et al. (2013). Morphology of two marine euplotids (Ciliophora: Euplotida), Aspidisca fusca kahl 1928 and A. hexeris quennerstedt 1869, with notes on their small subunit rRNA gene sequences. Eur. J. Protistol. 49, 634–643. doi: 10.1016/j.ejop.2013.04.005
Kahl A. (1928). Die infusorien (Ciliata) der oldesloer salzwasserstellen. Arch. Hydrobiol. 19, 50–123.
Kahl A. (1932). Urtiere oder Protozoa I: Wimpertiere oder ciliata (Infusoria) 3. spirotricha Vol. 25 (Tierwelt Dtl) (Jena: Gustav Fischer), 399–650.
Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Küppers G. C. (2020). A new species of Uronychia (Spirotrichea: Euplotida) from Argentina. Eur. J. Protistol. 75, 125706. doi: 10.1016/j.ejop.2020.125706
Lian C., Luo X., Fan X., Huang J., Yu Y., Bourland W. A., et al. (2018). Morphological and molecular redefinition of Euplotes platystoma dragesco & dragesco-kernéis 1986 and Aspidisca lynceus (Müller 1773) ehrenberg 1830, with reconsideration of a “Well-known” Euplotes ciliate, euplotes harpa stein 1859 (Ciliophora, euplotida). J. Eukaryot. Microbiol. 65, 531–543. doi: 10.1111/jeu.12499
Lian C., Wang Y., Jiang J., Yuan Q., Al-Farraj S. A., El-Serehy H. A., et al. (2021). Systematic positions and taxonomy of two new ciliates found in China: Euplotes tuffraui sp. nov. and E. shii sp. nov. (Alveolata, ciliophora, euplotida). Syst. Biol. 19, 359–374. doi: 10.1080/14772000.2020.1865472
Lian C., Wang Y., Li L., Al-Rasheid K. A. S., Jiang J., Song W. (2020). Taxonomy and SSU rDNA-based phylogeny of three new Euplotes species (Protozoa, ciliophora) from China seas. J. King. Saud. Univ. Sci. 32, 1286–1292. doi: 10.1016/j.jksus.2019.11.013
Li L., Shao C., Yi Z., Song W., Warren A., Al-Rasheid K. A. S., et al. (2008). Redescriptions and SSrRNA gene sequence analyses of two marine species of Aspidisca (Ciliophora, euplotida), with notes on the morphogenesis in A. orthopogon. Acta Protozool. 47, 83–94. doi: 10.1109/CSSE.2008.982
Li L., Zhang Q., Al-Rasheid K. A. S., Kwon C. B., Shin M. K. (2010). Morphological redescriptions of Aspidisca magna kahl 1932 and a. leptaspis fresenius 1865 (Ciliophora, euplotida), with notes on morphogenetic process in A. magna. Acta Protozool. 49, 327–337. doi: 10.4236/ojmh.2015.54008
Lynn D. H. (2008). The ciliated Protozoa: Characterization, classification, and guide to the literature. 3rd Edn (Dordrecht: Springer). doi: 10.1007/978-1-4020-8239-9
Medlin L., Elwood H. J., Stickel S., Sogin M. L. (1988). The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71, 491–499. doi: 10.1016/0378-1119(88)90066-2
Miller M. A., Pfeiffer W., Schwartz T. (2010). “Creating the CIPRES science gateway for inference of large phylogenetic trees,” in Paper presented at the 2010 gateway computing environments workshop (GCE) (New Orleans, LA: IEEE). doi: 10.1109/GCE.2010.5676129
Nylander J. A. A. (2004). MrModeltest 2.2. program distributed by the author (Uppsala, Sweden: Evolutionary Biology Centre, Uppsala University).
Page R. D. M. (1996). Treeview: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358. doi: 10.1093/bioinformatics/12.4.357
Pan X., Bourland W. A., Song W. (2013). Protargol synthesis: an in-house protocol. J. Eukaryot. Microbiol. 60, 609–614. doi: 10.1111/jeu.12067
Penn O., Privman E., Ashkenazy H., Landan G., Graur D., Pupko T. (2010). GUIDANCE: a web server for assessing alignment confidence scores. Nucleic Acids Res. 38, W23–W28. doi: 10.1093/nar/gkq443
Petz W., Song W., Wilbert N. (1995). Taxonomy and ecology of the ciliate fauna (Protozoa, ciliophora) in the endopagial and pelagial of the weddel Sea, Antarctica. Stapfia 40, 1–223.
Plough H. H. (1916). The genus Aspidisca ehrenberg. Trans. Am. Microsc. Soc 35, 233–244. doi: 10.2307/3221909
Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Serra V., Gammuto L., Nitla V., Castelli M., Lanzoni O., Sassera D., et al. (2020). Morphology, ultrastructure, genomics, and phylogeny of euplotes vanleeuwenhoeki sp. nov. and its ultra-reduced endosymbiont "Candidatus pinguicoccus supinus" sp. nov. Sci. Rep. 10, 1–27. doi: 10.1038/s41598-020-76348-z
Shen Z., Huang J., Lin X., Yi Z., Li J., Song W. (2010). Morphological and molecular characterization of Aspidisca hongkongensis spec. nov. (Ciliophora, euplotida) from the south China Sea. Eur. J. Protistol. 46, 204–211. doi: 10.1016/j.ejop.2010.01.004
Song W., Warren A., Hu X. (2009). Free-living ciliates in the bohai and yellow seas, China (Beijing: Science Press).
Song W., Wilbert N. (1997). Morphological investigation on some free living ciliates (Protozoa, ciliophora) from China Sea with description of a new hypotrichous genus, Hemigastrostyla nov. gen. Arch. Protistenkd. 148, 413–444. doi: 10.1016/S0003-9365(97)80020-6
Song W., Wilbert N. (2000). Ciliates from Antarctic Sea ice. Polar. Biol. 23, 212–222. doi: 10.1007/s003000050029
Song W., Wilbert N. (2002). Faunistic studies on marine ciliates from the Antarctic benthic area, including descriptions of one epizoic form, 6 new species and, 2 new genera (Protozoa: Ciliophora). Acta Protozool. 41, 23–61.
Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Syberg-Olsen M. J., Irwin N. A., Vannini C., Erra F., Di Giuseppe G., Boscaro V., et al. (2016). Biogeography and character evolution of the ciliate genus Euplotes (Spirotrichea, euplotia), with description of euplotes curdsi sp. nov. PloS One 11, e0165442. doi: 10.1371/journal.pone.0165442
Talavera G., Castresana J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577. doi: 10.1080/10635150701472164
Valbonesi A., Di Giuseppe G., Vallesi A., Luporini P. (2021). Two new species of Euplotes with cirrotype-9, euplotes foissneri sp. nov. and Euplotes warreni sp. nov. (Ciliophora, spirotrichea, euplotida), from the coasts of Patagonia: implications from their distant, early and late branching in the Euplotes phylogenetic tree. Int. J. Syst. Evol. Microbiol. 71, 4568. doi: 10.1099/ijsem.0.004568
Wang J., Zhang T., Li F., Warren A., Li Y., Shao C. (2021). A new hypotrich ciliate, oxytricha xianica sp. nov., with notes on the morphology and phylogeny of a Chinese population of Oxytricha auripunctata blatterer & foissne(Ciliophora, oxytrichidae). Mar. Life Sci. Technol. 3, 303–312. doi: 10.1007/s42995-020-00089-1
Warren A., Patterson D. J., Dunthorn M., Clamp J. C., Achilles-Day U. E. M., Aescht E., et al. (2017). Beyond the “Code”: a guide to the description and documentation of biodiversity in ciliated protists (Alveolata, ciliophora). J. Eukaryot. Microbiol. 64, 539–554. doi: 10.1111/jeu.12391
Wu I. C. H., Curds C. R. (1979). A guide to the species of the genus Aspidisca. Bull. Br. Mus.Nat. Hist. (Zool.). 36, 1–34.
Wu L., Li J., Warren A., Lin X. (2021). Taxonomy and systematics of a new pleurostomatid ciliate, Pseudolitonotus spirelis gen. et sp. n. (Protozoa, ciliophora, haptoria). Mar. Life Sci. Technol. 4, 31–41. doi: 10.1007/s42995-021-00123-w
Ye T., Zhao X., Chi Y., Zheng B., Zhang H., Huang J., et al. (2021). Morphology and molecular phylogeny of two marine folliculinid ciliates found in China (Ciliophora, heterotrichea). J. Ocean. Univ. China. 20, 959–972. doi: 10.1007/s11802-021-4965-z
Yi Z., Song W., Clamp J., Chen Z., Gao S., Zhang Q. (2009). Reconsideration of systematic relationships within the order euplotida (Protista, ciliophora) using new sequences of the gene coding for small-subunit rRNA and testing the use of combined data sets to construct phylogenies of the Diophrys-complex. Mol. Phylogenet. Evol. 50, 599–607. doi: 10.1016/j.ympev.2008.12.006
Zhang C., Huang J., Ye T., Lu B., Chen X. (2020). The morphology and phylogeny of three Diophrys ciliates collected from the subtropical waters of China, including a new species (Ciliophora; euplotia). J. Ocean. Univ. China 19, 975–987. doi: 10.1007/s11802-020-4536-8
Keywords: Aspidisca, new species, morphology, SSU rRNA gene sequence, phylogeny
Citation: Zheng B, Zhao X, Ye T, Huang J, Vallesi A, Jiang Y and Chen X (2022) Morphology and molecular phylogeny of two new Aspidisca species (Ciliophora, Spirotrichea, Euplotida) collected from subtropical coastal waters in China. Front. Mar. Sci. 9:970692. doi: 10.3389/fmars.2022.970692
Received: 16 June 2022; Accepted: 18 July 2022;
Published: 08 August 2022.
Edited by:
Xiaofeng Lin, Xiamen University, ChinaReviewed by:
Jiamei Jiang, Shanghai Ocean University, ChinaKesava Priyan Ramasamy, Umeå University, Sweden
Copyright © 2022 Zheng, Zhao, Ye, Huang, Vallesi, Jiang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaohan Jiang, jiangyaohan1993@163.com; Xiangrui Chen, chenxiangrui@nbu.edu.cn
†These authors have contributed equally to this work
 Bohan Zheng
Bohan Zheng Xuetong Zhao1†
Xuetong Zhao1†  Adriana Vallesi
Adriana Vallesi Xiangrui Chen
Xiangrui Chen