- 1Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China
- 2College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing, China
- 3School of Life Sciences, Jinggangshan University, Ji’an, China
There is increasing evidence that species diversity is underestimated in the current taxonomy of widespread freshwater fishes. The bagrid species T. albomarginatus s.l. is mainly distributed in the lowlands of South China, as currently identified. A total of 40 localities (including the type locality), which covers most of its known range, were sampled. Molecular phylogenetic analyses based on concatenated mtDNA and nuclear genes recover nine highly supported lineages clustering into eight geographic populations. The integration of molecular evidence, morphological data, and geographic distribution demonstrates the delineation of T. albomarginatus s.l. as eight putative species. Four species, namely, T. albomarginatus, T. lani, T. analis, and T. zhangfei sp. nov. and the T. similis complex are taxonomically recognized herein. Moreover, T. zhangfei sp. nov. comprises two genetically distinct lineages with no morphological and geographical difference. This study also reveals aspects of estimation of divergence time, distribution, and ecological adaption within the T. albomarginatus group. The unraveling of the hidden species diversity of this lowland bagrid fish highlights the need for not only the molecular scrutiny of widely distributed species of South China but also the adjustment of current biodiversity conservation strategies to protect the largely overlooked diversity of fishes from low-elevation rapids.
Introduction
Freshwater ecosystems not only harbor incredible amounts of biodiversity but also provide significant ecosystem services for mankind (Harrison, et al., 2016). However, the threat under which freshwater ecosystems have been put is far greater than that of territorial or marine ecosystems (Darwall, et al., 2018). As an essential composition of this ecosystem, freshwater fishes are currently confronted with severe threats from anthropogenic perturbations (Schmidt et al., 2019). It is therefore urgently needed to take protection actions for freshwater ecosystems in general and for freshwater fishes in particular (Gascón et al., 2008; Rojas et al., 2020). Insufficient basic knowledge regarding species diversity is frequently one of primary impediments for freshwater fish conservation (Darwall et al., 2018). Protection strategies for either freshwater ecosystems or freshwater fishes can become misdirected due to failure to detect the true species diversity, particularly hidden diversity of widespread species. Increasing evidence has shown that widespread species, which are often recognized as a single species owing to very similar or indistinguishable morphology, can comprise multiple species (Das, 2007).
The delineation of hidden diversity within widespread species has the great potential to further our understanding of macroecology, evolutionary processes, and conservation biology (Brickford et al., 2007). The practice of integrative taxonomy can yield many more putative species than traditional taxonomy, which would result in improved biodiversity assessments allowing better modeling of macroecological processes and increased resolution of spatial heterogeneity (Eme et al., 2017). The detection of putative species also has evolutionary significance as they share similar morphology with low rates of phenotypic variation (Tan et al., 2010). Additionally, the average species distribution range decreases drastically with the growing number of species. Species with small range sizes are much more vulnerable to anthropogenic or natural factors than broadly distributed species (Delic et al., 2017). Neglect of the vulnerability of putative species can lead to underestimating threats to biodiversity.
Although the detection of species diversity poses a big challenge for traditional taxonomy, the process of species exploration has been greatly accelerated by the use of current molecular techniques (Kekkonen and Hebert, 2014). Generally, phylogenetic lineages recovered in most cases of molecular analysis can merit their specific status with more or less morphological and/or ecological distinctness (Ozkilinc and Sevinc, 2018). The last decade has witnessed the utilization of molecular technology to unveil the genetic structure differentiations of widely distributed Chinese freshwater fishes, including Hemicculter leuciclus (Chen et al., 2017), Garra orientalis (Yang et al., 2016), Squalidus argentatus (Yang et al., 2013), Hemibagrus macropterus (Lei et al., 2009), and Opsariichthys bidens (Perdices et al., 2005). Unfortunately, few studies incorporated molecular datasets with morphological evidence to argue for the specific status of recovered genetic lineages. Moreover, it has been revealed that results only from molecular approaches should be utilized with caution for species delineation (Forister et al., 2008). The use of different molecular markers and species delineation methods can lead to conflicting results about the number of delineated species. Seeking congruence between molecular and non-molecular data using integrative taxonomy is currently becoming the principal methodology in species exploration (Welton et al., 2014).
South China lies within the Asian subtropical region with an affluent diversity of freshwater fish species (Du et al., 2021). In stark contrast to the high amount of conservation attention currently given to the highlands of South China, including the upper Yangtze River (= Chang-Jiang in Chinese) and upper Pearl River (=Zhu-Jiang in Chinese) basins, which have high numbers of endemic and threatened fishes (Dao et al., 2020), little or no efforts have been made to protect the non-charismatic fish species of the lowlands of South China such as the mid-lower Chang-Jiang and Zhu-Jiang basins, likely due to a lower level of endemism. However, several widespread freshwater fishes from these lowland areas have been shown to represent multiple distinct genetic lineages by using phylogenetic analysis (Perdices et al., 2005; Yang et al., 2016; Chen et al., 2017), therefore justifying a need to reassess the morphologically based widespread species under a molecular scrutiny. This is the case for the currently identified widespread bagrid species Tachysurus albomarginatus from South China.
Tachysurus albomarginatus was originally described by Rendahl (1928) on the basis of five 49.0–90.0-mm SL specimens caught from Tang-tu-hsien [now Dangtu County, in the lower Chang-Jiang basin], Anhui Province, South China. The genus Tachysurus La Cepède, 1803, with which both Pseudobagrus and Peltobagrus were synonymized (Ng and Kottelat, 2007), is an endemic East Asian group of small or medium-sized species adapted to the fast-flowing waters of mountain streams or main channels of rivers (Ng and Freyhof, 2007). Although there is ongoing controversy over the specific status of T. albomarginatus, the majority of authors since Rendahl have regarded it valid (Chen, 1984; Gao, 1990). In Chinese literature, T. albomarginatus is a species widely distributed mainly in the lowlands of South China such as the mid-lower Chang-Jiang, middle Zhu-Jiang, Qiantang-Jiang, and Min-Jiang basins, and all other coastal rivers of Zhejiang and Fujian provinces, with its northern extent at the lower Huang-He and Huai-He basins (note: the suffixes -Jiang and -He in Mandarin Chinese mean river) (Gao, 1990; Cheng and Zhou, 1997). The present identification of this lotic bagrid species is far from satisfactory. Detailed comparisons of available specimens, coupled with a brief review of literature records, showed that T. albomarginatus, as conventionally defined, contains multiple species deserving specific recognition (Cheng et al., 2021). A thorough taxonomic revision of this species based on more sufficient sampling is urgently needed.
The objective of this study is to explore the possible hidden species diversity within Tachysurus albomarginatus s.l. utilizing integrative approaches. Molecular phylogenetic relationships among intraspecific populations were inferred based on the concatenated mitochondrial and nuclear genes of specimens extensively sampled throughout the majority of its known range. Morphometric measurements and meristic counts were analyzed to test for morphological differentiations among genetic clustering groups. Species delineation within T. albomarginatus s.l. was predicated on the congruence between morphological and genetic datasets. The findings of all the above analyses highlight a need for taxonomic reevaluation of currently identified widely distributed species and reconsideration of current protection strategies of freshwater fishes from South China, particularly those found in relatively lower elevations.
Materials and Methods
Taxon Sampling, Specimen Collection, and Preservation
Forty sampling locations throughout most of the historically known distribution of Tachysurus albomarginatus s.l. were selected, spanning 14 river basins (Figure 1). Sampling localities were numbered from 1 to 40. A total of 418 individuals of the currently recognized T. albormaginatus were collected. Among them, 127 individuals were used for molecular analysis, 297 for morphometric analysis, and 207 for meristic counts (see Table 1). Sequences from samples of other 16 congeneric species were also used in the phylogenetic analysis (Table 2), and Hemibagrus macropterus was selected as the outgroup.
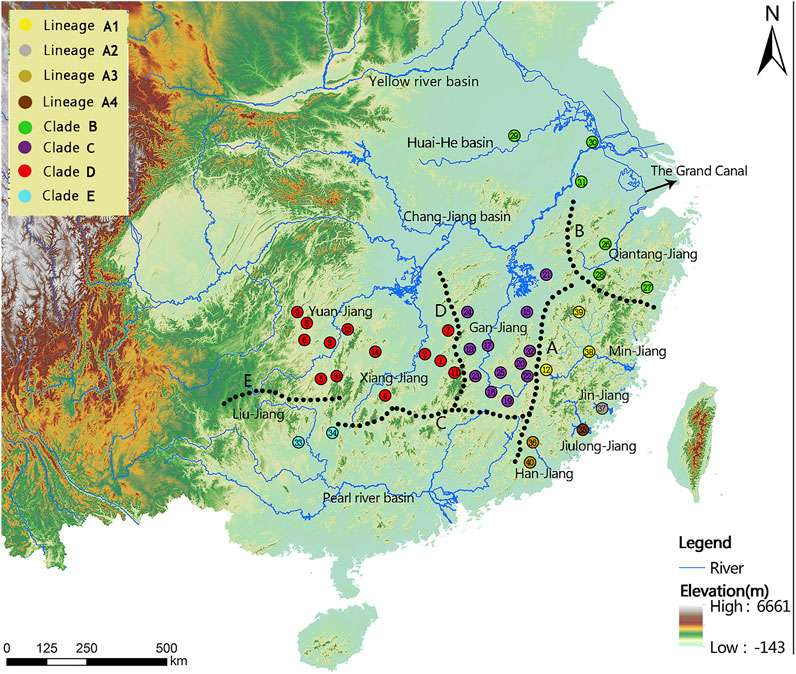
FIGURE 1. Sampling localities (1-40) and geographical barriers (A–E) within the T. albomarginatus species group. Distinct colors denote the same lineage membership as used in Figures 2–6. A, Wuyi Mountains; B, Huangshan-Tianmushan Mountains; C, Nanling Mountains; D, Luoxiao-Mufu Mountains; E, Miaoling Mountains.
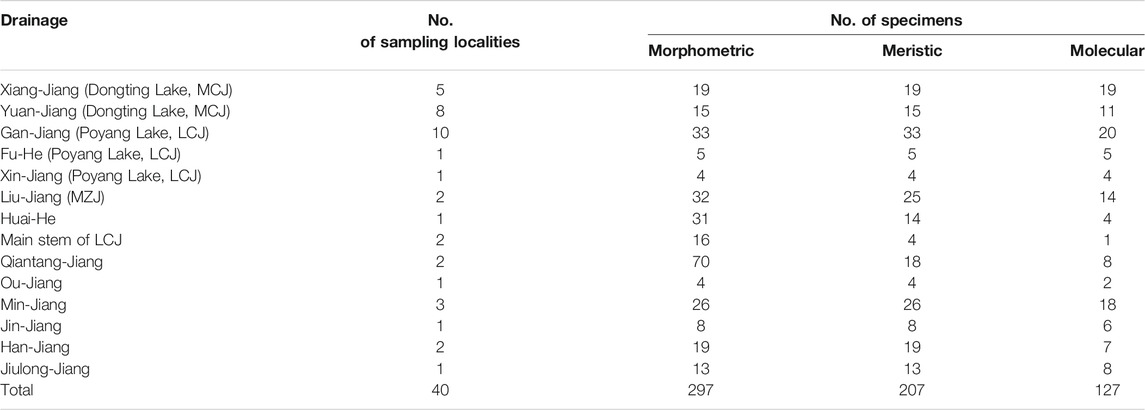
TABLE 1. Detailed sampling drainage with locality and specimen numbers (MCJ: middle Chang-Jiang basin; LCJ: lower Chang-Jiang basin; MZJ: middle Zhu-Jiang basin).
DNA Extraction, Amplification, and Sequencing Analyses
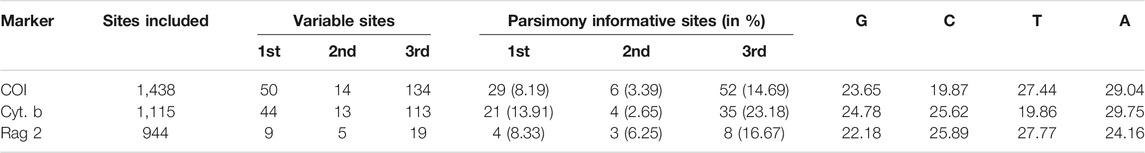
Genomic DNA was extracted from alcohol-preserved fin clips using the DNeasy Tissue Kit (Qiagen, Beijing, China). Two mitochondrial genes [cytochrome b (Cyt. b) and cytochrome c oxidase subunit I (COI)] and one nuclear gene [recombination activating 2 (RAG2)] were amplified using polymerase chain reaction (PCR). The information for the primers used in this study is shown in Table 3. The following thermal cycling profiles were adopted: 95°C predenaturing (5 min), 95°C dematuring (30 s), 58°C for RAG2 or 52°C for Cyt. b and COI annealing (45 s), 72°C extension (90 s), for 35 cycles, and 72°C final extension (8 min). Amplified products were subsequently purified and utilized for direct cycle sequencing by Tianyi Huiyaun Sequencing Company. Sequence data were archived in the public domain database GenBank (Table 2). Multiple alignments were prepared utilizing MEGA 7.0 for all sequences, based on the amino acid sequences with the program MUSCLE (Edgar, 2004) with the default setting.
Phylogenetic Analyses
COI, Cyt. b, and RAG2 genes of each analyzed specimen were concatenated to form a dataset for phylogenetic tree reconstructions with Bayesian inference (BI) and Maximum likelihood (ML). The general time-reversible model with invariant sites and a gamma distribution variation across sites (GTR + G) was selected as the best-fitting model for COI, Cyt. b, and RAG2 in Jmodeltest2 (Darriba et al., 2012) based on Akaike informative criterion (Akaike, 1974). MrBayes 3.1.2 (Ronquist and Huelsenbeck, 2003) was applied to calculate BI. Two independent parallel Markov chain Monte Carlo runs with four chains for 50,000,000 generations were performed, sampling every 1,000 generations and discarding the first 25% samples as burn-in. Sufficient convergence of the Markov chain Monte Carlo (MCMC) results was verified based on summary statistics in MrBayes v3.2.6. A maximum likelihood tree of the concatenated dataset was generated with RAxML GUI v.1.5b, applying GTR + G to both COI and Cyt. b (Silvestro & Michalak, 2012). Bootstrap values (BS) of nodes were estimated by 1,000 bootstrap replicates. All trees were rooted by Hemibagrus macropterus and visualized in FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
Haplotype networks were constructed for the concatenated mtDNA sequence datasets to better visualize relationships in multifurcations and reticulations (Posada and Crandall, 2001). A maximum parsimony method, tcs v.1.23 (Clement et al., 2000), was used to draw an unrooted network to evaluate the haplotype relationships for the mtDNA sequences, with 95% parsimoniously plausible branch connections. The genetic distance based on the K2P (Kimura two-parameter) model (Kimura, 1980) was calculated with MEGA 7.0. DNASP v5 was used to filter the haplotype (Librado and Rozas, 2009).
Species Delineation
Two independent methods relying on different operational criteria were applied to infer molecular species delineation for T. albomarginatus s.l.: the automatic barcode gap detection (ABGD) and the Poisson tree process (PTP) model. The automatic barcode gap discovery (ABGD) is a barcode species delineation method aiming to establish a minimum gap that probably corresponds to the threshold between interspecific and intraspecific processes (Puillandre et al., 2012). The major advantage of the ABGD method, when compared to the other barcode species delineation ones, is that the inference of the limit between interspecific and intraspecific processes is recursively applied to previously obtained groups to get finer partitions until there is no further partitioning, thus allowing a more refined search. Basically, the ABGD analysis indicates the number of groups (species) estimated relative to a large spectrum of p values (prior intraspecific values) in which a 0.1 value assumes a maximum intraspecific variability, suggesting that all sequences belong to only one species, whereas a 0.001 value assumes a very small intraspecific variability suggesting that each distinct haplotype represents a different species. After performing the ABGD analysis, additional molecular, morphological, or ecological characters are needed to infer the correct number of species. The ABGD analysis was run on the ABGD server website (https://bioinfo.mnhn.fr/abi/public/abgd/abgdweb.html).
The PTP is a tree-based species delineation method under the assumption that the intraspecific substitution number is significantly less than the interspecific substitution number (Kapli et al., 2017). The Bayesian (PTP) and maximum likelihood (mPTP) implementations from concatenated data were used in our study. The branch lengths of the phylogenetic input tree represent the number of substitutions. Unrooted phylogenetic trees, without outgroup, created by MrBayes and RAxML were uploaded to the online server of PTP and mPTP (http://species.h-its.org/ptp/).
Divergence Time Estimation
We estimated the divergence time of T. albomarginatus s.l. using the birth/death speciation process model in Beast v1.10.4 with concatenated data of Cyt. b and COI sequences (Drummond and Rambaut, 2007). Best-fit models of Cyt. b and COI were applied in estimation of divergence time. We adopted a substitution rate of 0.18–0.30% per million years for cyt. b, which is proposed by Peng et al. (2002) for the East Asian bagrid catfishes. The uncorrelated log-normal relaxed clock was applied to Cyt. b and COI. The mean and standard deviation of the Cyt. b substitution rate, i.e., its ucldMean and ucldStdev, were fixed to 0.0024 and 0.136, setting a 95% highest posterior density interval (HPD95) to 0.18–0.30% per million years for substitution rate. The mean rate and standard deviation of COI were estimated in the MCMC run. A single MCMC run was performed with 200,000,000 generations, sampling every 1,000 generations. Sufficient convergence and burn-in of runs were checked in Tracer v1.7 (http://tree.bio.ed.ac.uk/software/tracer) to ascertain that all effective sampling size (ESS) values are above 200. The first 10% of samples were discarded as burn-in in TreeAnnotator v1.10.4 in the Beast package (Drummond and Rambaut, 2007). A consensus tree was generated with maximum clade credibility. The consensus tree was visualized in FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
Morphological Analysis
Twenty-six morphometric measurements utilized in this study are presented in Table 6. These measurements were made on the left side of each individual wherever possible. Following the methods of Cheng et al. (2008), data were taken point to point with digital calipers linked to a data recording computer and recorded to the nearest 0.1 mm. Morphometric measurements were subject to principal component analysis (PCA) in order to check external morphological differentiation and explore the relative contributions of specific variables to morphological differences in the target species.
PCA was run with SPSS 16 (SPSS, Chicago, IL, USA). Prior to the analysis, the method of Reist (1985) was used to normalize all morphometric measurements except standard length to eliminate the influence of allometry of body parts and sample size on the morphometric data. The formula of the corrected measurements was given as follows: Mtrans = log M-b (log BL-log BLm) where Mtrans is the size-transformed measurement for each individual; M is the original unadjusted measurement; b is the allometric coefficient that was calculated as the slope of log M against log BL; BL is the standard length of each individual; and BLm is the overall mean standard length of one population while log is the base 10 logarithm. All measurements except standard length were transformed separately utilizing the regression slope and common overall mean body length. The PCA loadings are presented in Table 6.
The counts of vertebrae were taken from photographs of Micro-CT (X-ray-based micro-computed tomography) scanning to explore internal morphology. Micro-CT scanning was made in a Siemens SOMATOM Definition X-ray machine (120 kV, 400 mA, and 0.4 mm slice thickness and voxel dimensions 0.16 mm × 0.16 mm×0.16 mm). Micro-CT data were imported into the Mimics medical imaging software (Materialise N.V., Leuven, Belgium), and a 3D digital reconstruction of each block was created. The individual reconstructions were tidied, smoothed, and stitched together utilizing Adobe Photoshop (Adobe Systems Inc., Mountain View, CA, USA).
Results
Sequence Characteristics
The sequence information for the collection used in the molecular phylogenetic analysis is shown in Table 4. A total of 3,497 bp nucleotides were sequenced for one nuclear and two mitochondrial genes: RAG 2 (944 bp), COI (1,438 bp), and Cyt. b (1,115 bp). The nucleotide sequences of these three protein-coding genes code for 314, 479, and 371 amino acids, respectively. For the whole dataset, 401 characters are variable and 162 characters are parsimony informative. The variable and parsimony sites of the RAG2 gene are both fewer than those of CO1 and Cyt. b, thus suggesting the former as being less informative when compared to the latter two mitochondrial genes.
Phylogenetic Analysis and Genetic Distances
Phylogenetic trees for T. albomarginatus s.l. are inferred from the combined mtDNA (COI and Cyt. b) and nuclear (RAG2) genes using two different inference approaches (ML and BI). Analyses based on these two approaches results in the same general topology (Figure 2). All samples here recognized as T. albomarginatus s.l. constitute a monophyletic assemblage strongly supported by a 100% bootstrap value (hereafter bv) and posterior probability (hereafter pp) and cluster into five clades (A, B, C, D, and E) well supported by 100% bv and 100% pp. While Clades B, C, and E include a single lineage (hereafter also called Lineage B, C, or E for convenience of comparison), Clades A and D comprise four and two lineages, respectively. Clade E is the earliest diverging lineage including samples from the Liu-Jiang of the Zhu-Jiang basin. It is supported by 96% bv and 72% pp to be sister to the group formed by the remaining four clades where Clade D is the basal lineage and the weakly supported (less than 50% bv and 81% pp). Paired clades (B and C) receive a strong support of 100% bv and 91% pp to be sister to Clade A. Clade D is constituted by samples from the Yuan-Jiang and Xiang-Jiang, two affluents of Lake Dongting linked to the middle Chang-Jiang, and comprises two sister lineages (D1 and D2), sympatrically occurring or having a entirely overlapping distribution. Clade C is made up of samples from the Xin-Jiang, Fu-He, and Gan-Jiang, three affluents of Lake Poyang connected with the mainstem of the lower Chang-Jiang. Clade B is composed of samples from the lower Huai-He basin, the main stream of the lower Chang-Jiang, and coastal river basins of Zhejiang Province such as the Qiantang-Jiang and Ou-Jiang. There are four lineages (A1-A4) clustered within Clade A where Lineage A4 is sister to the remaining three ones constituting a polytomy. These lineages are each weakly supported but exhibited a significant regional clustering as they correspond to four coastal river basins of Fujian Province. Lineages A1-A4 are each found only in the Min-Jiang, Jin-Jiang, Han-Jiang, and Jiulong-Jiang basins, respectively.
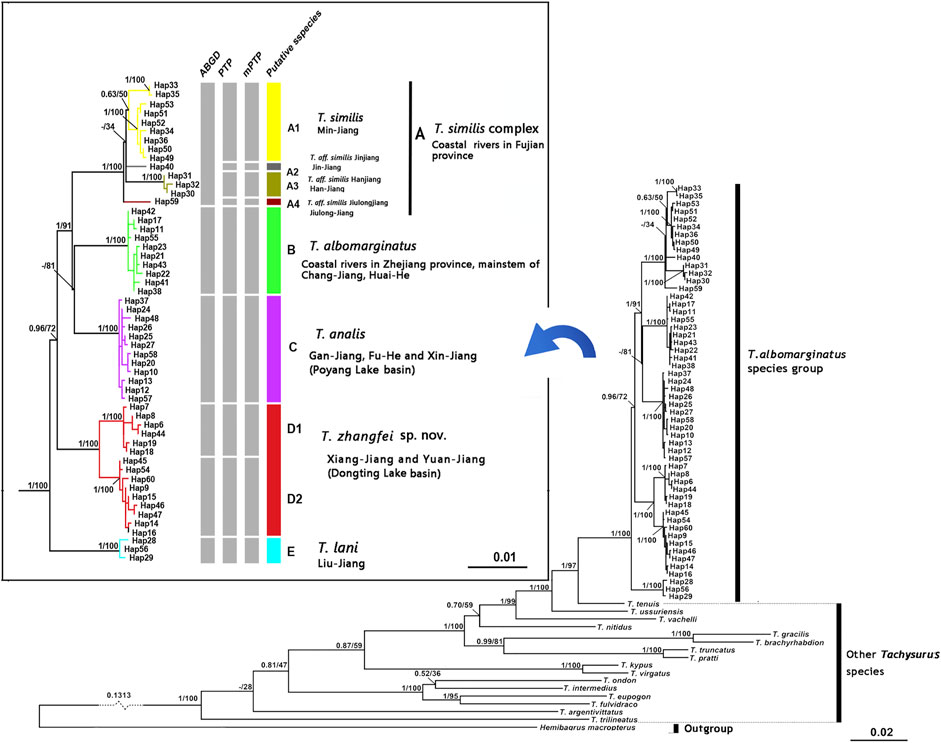
FIGURE 2. Bayesian tree based on combined genes for the T. albomarginatus species group. Values on the left of the nodes indicate posterior probabilities and bootstrap supports of Bayesian inference/maximum likelihood for major lineages. The results of species delimitation are shown on the right.
The paired Clades A and B and four lineages within Clade A receive a low nodal support and have short branch length in the molecular phylogenetic tree of the T. albormarginatus s.l. species group (Figure 2); however, the median-joining network analysis corroborates the split of nine lineages (A1, A2, A3, A4, B, C, D1, D2, and E) recovered here and no haplotype is found to be shared among these lineages (Figure 3). Besides, four lineages (A1-A4) within Clade A form network-like patterns instead of dichotomic cladogenesis in the haplotype network analysis.
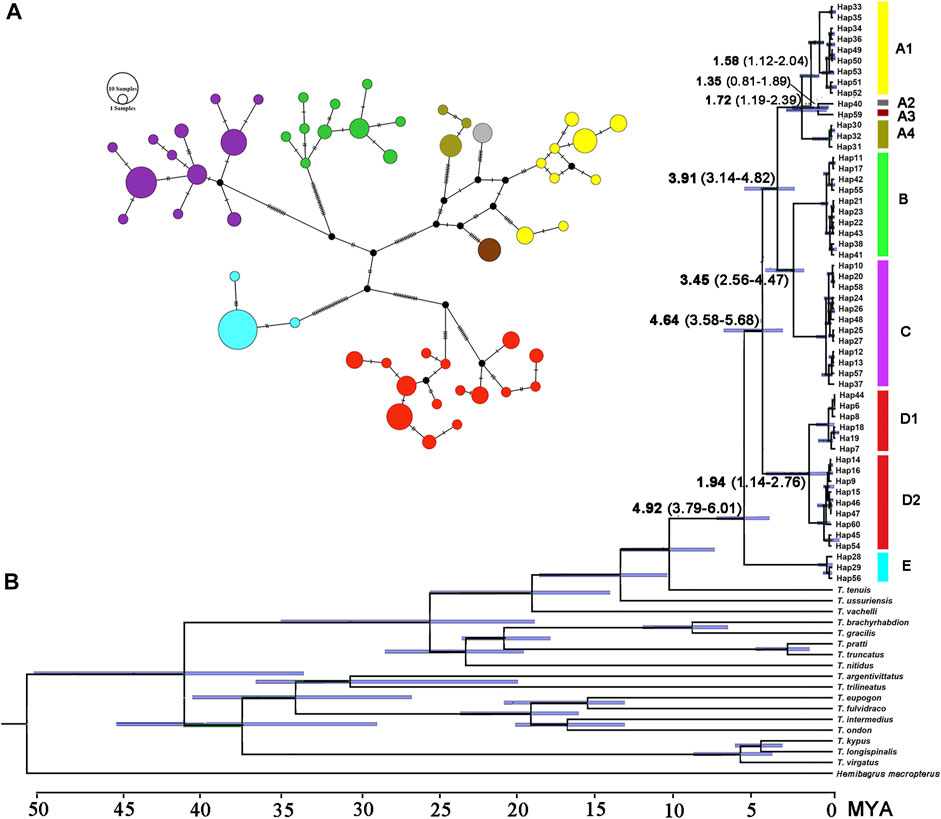
FIGURE 3. (A) Median-joining network (each circle represents a haplotype proportional to its frequency colored by the genetic split in the phylogenetic tree) and (B) the chronogram (BEAST) of the main divergence events in T. albomarginatus s.l. inferred for the concatenated Cyt. b and COI data.
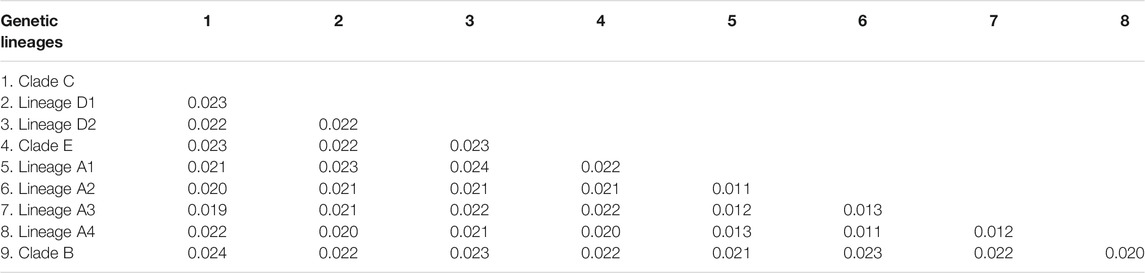
The estimated K2P genetic distances of the Cyt. b gene between T. albomarginatus s.l. and congeneric species range from 5.6 to 12.5% and those between paired lineages within T. albomarginatus s.l. range from 1.1 to 2.4% (Table 5). The mean genetic distance among these lineages is 1.7%, and the range of intraspecific genetic distances within each lineage is 0.1–0.2%. Four allopatric lineages (Lineages A1 to A4) from the coastal rivers of Fujian Province exhibit K2P genetic distances from 1.1 to 1.3%, while sympatric paired Lineages D1 and D2 from the Xiang-Jiang and Yuan-Jiang have a 2.2% genetic distance. The K2P distances of COI among/within nine lineages are at the range of 1.2–2.5%/0.1–0.2%. There is relatively less genetic divergence for nuclear genes than for mitochondrial genes, and K2P distances of RAG2 among nine lineages are ranged at 0.1–0.3%.
Divergence Time Estimate
The estimated time, with a 95% highest posterior density (HPD), is shown in Figure 3B. The most recent common ancestor of the T. albomarginatus group was estimated to be around 4.92 Ma (95% HPD: 3.79–6.01) (Figure 3B). Following the divergence of Clade E, Clade D branched off from the remaining clades 4.64 Ma (95% HPD: 3.56–5.68). The time that Clade A separated from the paired Clades B and C was about 3.91 Ma (95% HPD: 3.14–4.82). The diverging time between Clades B and C was inferred to be about 3.45 Ma (95% HPD: 2.56–4.47). Within Clade A, Lineage A4 deeply split 1.72 Ma (95% HPD: 1.19–2.39); the subsequent divergence of Lineage A1 occurred about 1.58 Ma (95% HPD: 1.12–2.04), Lineage A2 and A3 diverged 1.35 Ma (95% HPD: 0.81–1.69).
Molecular Species Delineation
The ABGD analysis supports six molecular operational taxonomic units (MOTUs) at a prior maximal distance of 0.0100, and these six MOTUs correspond to four clades (A, B, C, and E) and two lineages (D1 and D2), recovered in the molecular phylogenetic analysis based on the combined mtDNA and nuclear genes (Figure 2). Concurrently, the PTP analysis identifies nine MOTUs with the posterior probabilities (higher than >0.70) of 0.90, 0.88, 0.99, 0.99, 0.91, 0.96, 0.89, 0.90, and 0.92; these MOTUs correspond to three clades (B, C, E) and six lineages (A1, A2, A3, A4, D1, and D2). The same result is repeated in the mPTP analysis where the recognition of these MOTUs is supported with the posterior probabilities of 0.91, 0.92, 0.99, 0.95, 0.98, 0.93, 0.92, 0.90, and 0.88, respectively. The difference in species delineation between these two methods is that four lineages (A1-A4), clustered within Clade A from four coastal river basins of Fujian Province, are delimited by the ABGD method as only one MOTU and by the PTP method as four MOTUs.
Morphological Distinctions
The nine distinct lineages detected in the molecular phylogenetic analysis correspond to eight geographical populations (Figure 2). Paired lineages D1 and D2 are coexisting and form a single geographical population. Each of the remaining seven lineages (A1, A2, A3, A4, B, C, E) represents its own geographical population. The PCA performed for 26 morphometric measurements (Table 6) show, to some extent, variations among these eight geographic populations. The scatter plot of PC2 versus PC3 calculated for eight populations indicates that they cluster into four groups (Figure 4A). Clade B and Lineage A2 each form a distinct group completely separating from all other groups. Clades C, D, and E are grouped together, largely overlapping with each other, and so does Lineages A1, A3, and A4. Lineages A1 and A4 cluster together or are unseparated but slightly overlap with Lineage A3. The sympatric paired Lineages D1 and D2 are not distinguishable in either morphometric data or meristic counts (Supplementary Figure S1). The scatterplot of PC1 vs. PC2 shows the similar pattern with that of PC2 vs. PC3 except that Lineages A1-A4 heavily overlap with each other (Figure 4B).
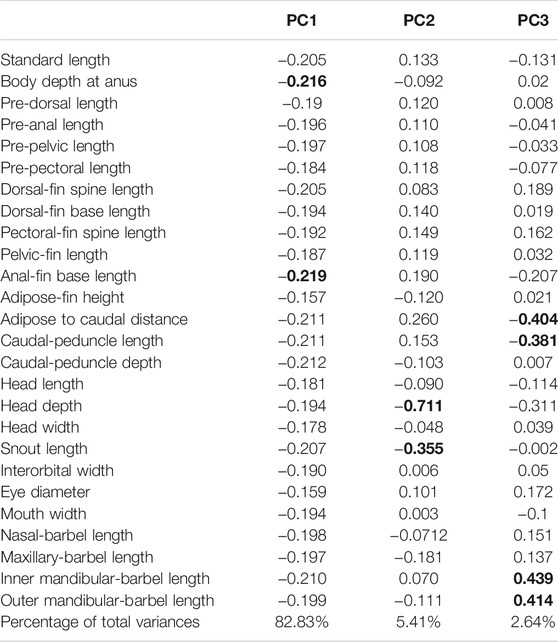
TABLE 6. Variable loadings in the first to third axes of the principal component analysis among eight allopatric populations within T. albomarginatus s.l.
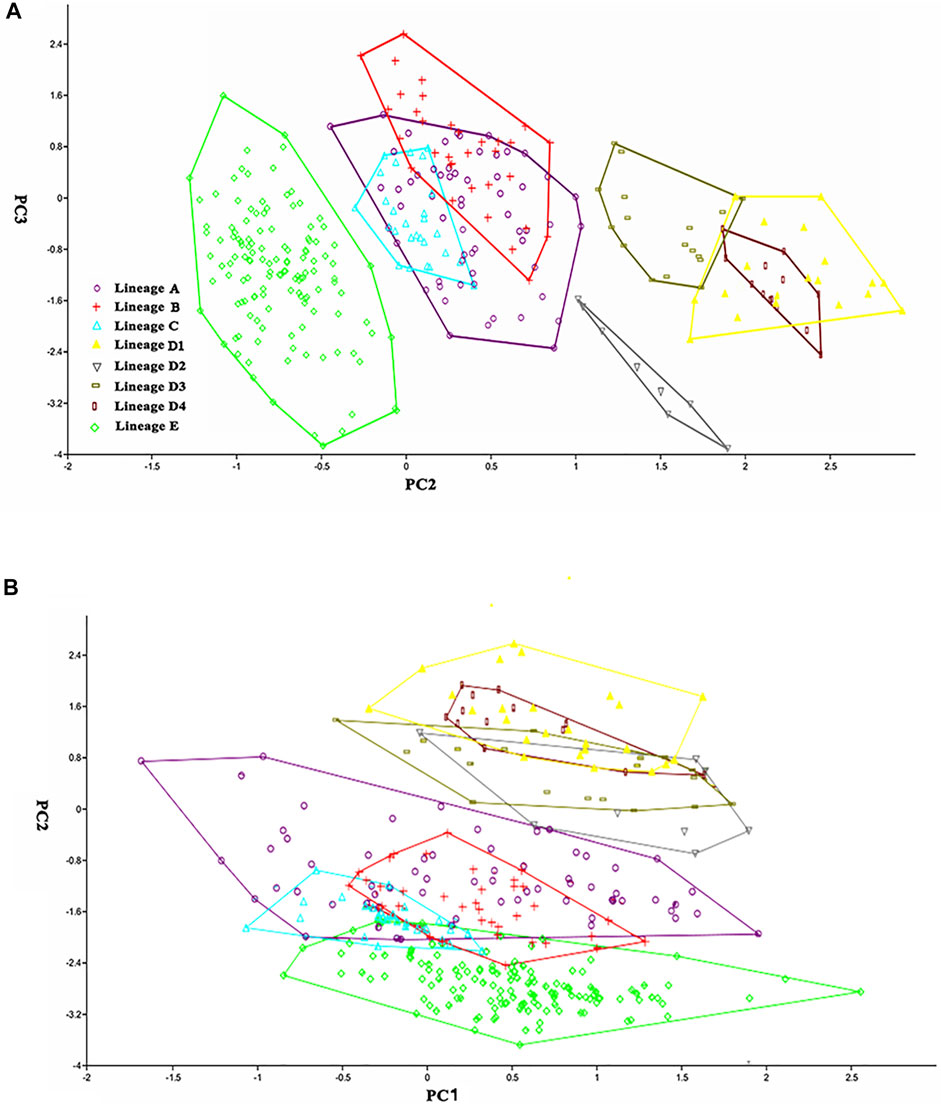
FIGURE 4. Scatter plot of (A) PC2 against PC3 and (B) PC1 against PC2 extracted from morphometric data for eight geographic populations.
There are two morphometric measurements (anal-fin base length and body depth at anus) with main loadings on the first axis (PC1), three (adipose to caudal distance, head depth, and snout length) on the second axis (PC2), and two (inner and outer mandibular-barbel lengths) on the third axis (PC3) (Table 6). Stable differences in the head depth, outer mandibular-barbel length, and body depth at the anus are found among these lineages (Figure 6). Counts of vertebrae and anal-fin rays are found to distinguish among them (Figure 4). These eight geographical populations can be clearly distinguishable through the combination of morphometric measurements and meristic counts.
Clade C and Lineage A1 differ from all other lineages in having more vertebrae numbers (47–49 vs. 39–46; Figure 5A), but both differ from each other in the head depth (58.4–76.1% HL vs. 43.6–55.6; Figure 6A). Clade D is separated from all other lineages except Lineages A2 and A3 in having fewer vertebrae (39–43 vs. 44–49; Figure 5A). It has a slight overlap in this character (39–43 vs. 43–45; Figure 5A) with Lineages A2 and A3 but differs from the former in having fewer anal-fin rays (15–18 vs. 19–21; Figure 5B) and from the latter in possessing longer outer mandibular barbels (length 24.4–36.8% HL vs. 21.1–23.9; Figure 6B). Clades B and E differ from Lineages A2-A4 in having a deeper head (depth 67.7–89.1% of HL vs. 43.16–66.4), and the head depth is greater in Clade B than in Clade E (Figure 6C). Lineage A2 differs from Lineages A3 and A4 in having shorter mandibular barbels (length 21.1–23.9% of HL vs. 23.53–34.0) and a deeper head (depth 58.6–66.4% of HL vs. 43.1–55.7) (Figures 6C,D), from Clade E in having a deeper body (depth at anus 11.1–14.2% of SL vs. 15.4–21.2) (Figure 6E). Mandibular barbels are longer in Lineage A3 than in Lineage A4 (Figure 6D).
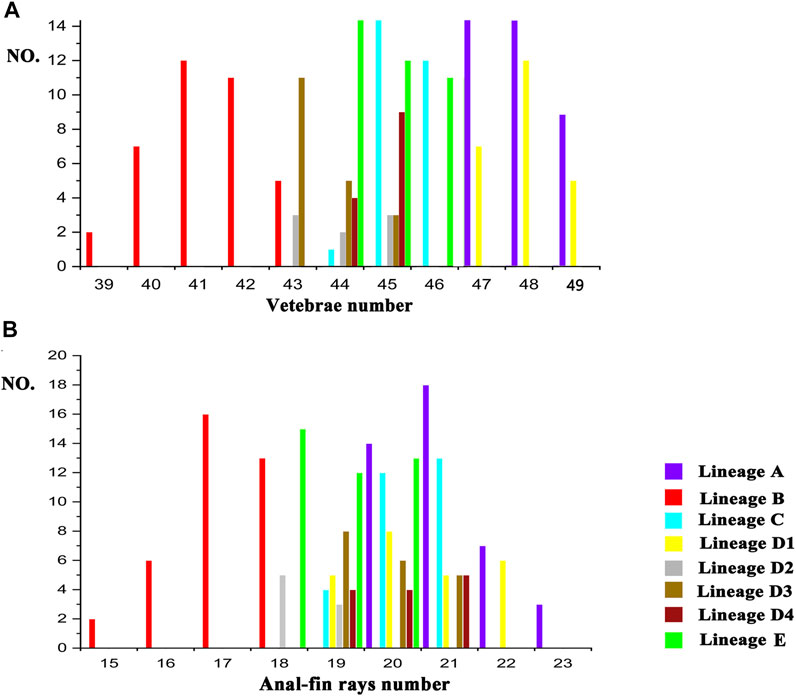
FIGURE 5. Meristic counts difference among eight geographic populations in vertebrae number (A) and anal-fin ray number (B).
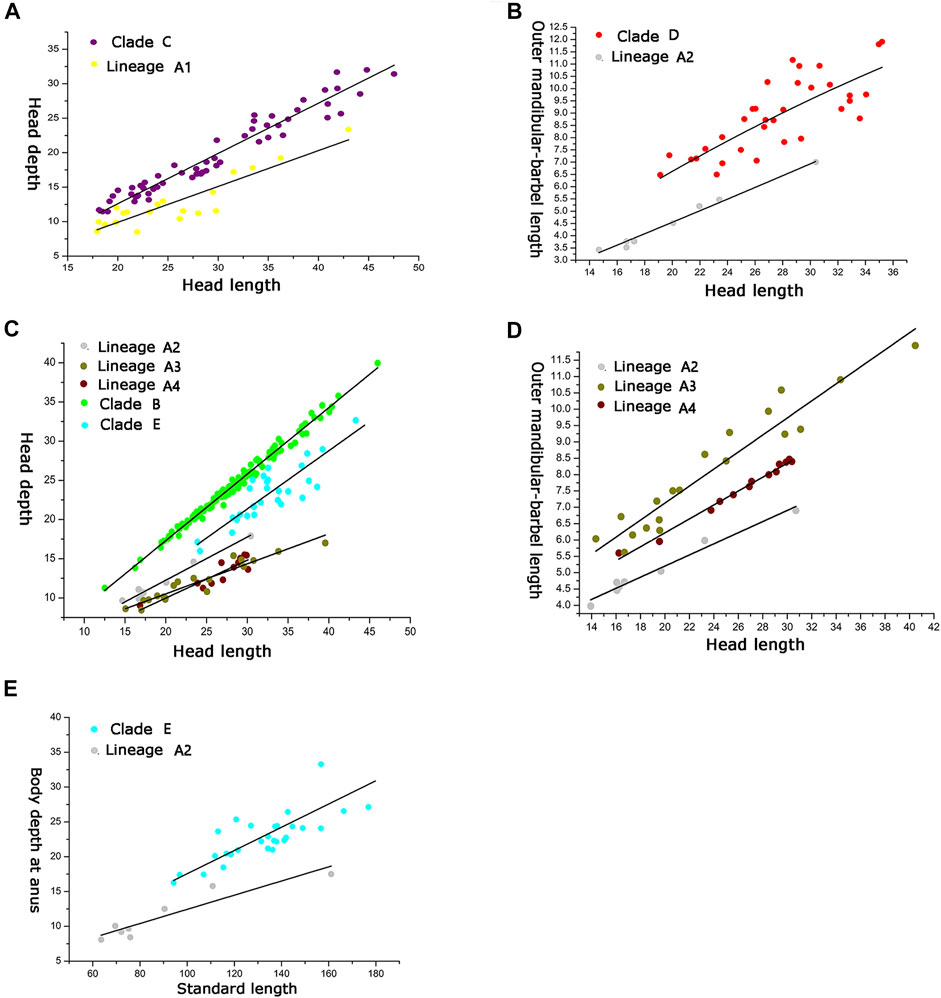
FIGURE 6. Relation between (A) head depth and HL for Clade C and Lineage A1; relation between (B) outer mandibular barbel length and HL for Clade D and Lineage A2; relation among (C) head depth and HL for Clades A and B, Lineages A2, A3, and A4; relation among (D) outer mandibular barbel length and HL for Lineages A2, A3, and A4; relation between (E) body depth at anus and SL for Clade E and Lineage A2.
Discussion
Hidden Diversity
The hidden species diversity of the morphology-based species T. albomarginatus s.l. is unveiled in this study. Nine distinct lineages are recovered in the phylogenetic analysis based on combined mtDNA (COI and Cyt. b) and nuclear (RAG2) genes, and the use of different molecular species delineation methods results in the detection of six or nine MOTUs (Figure 2). The mismatch in molecular species delineation utilizing these two methods (ABGD and PTP analysis) arises from the delimitation of Clade A as one or four MOTUs. It is not surprising that the ABGD method has a relatively conservative species delineation for the clade, given the small genetic divergences among constituent lineages (A1-A4). Previous investigations have demonstrated that the ABGD analysis, which is based only on nucleotide divergence to delineate species (Laris et al., 2015), fails to distinguish sister species that are easily recognizable from morphology (Lessios, 2008; Silva et al., 2018). In contrast, the PTP analysis requires neither an ultrametric input tree nor a sequence similarity threshold as input and thus outperformed the other methods when genetic divergences between species are small (Kapli et al., 2017). In the case of the T. albomarginatus s.l., Lineages A1-A4 are delineated by this method as four molecular operational taxonomic units (MOTUs), despite their small genetic divergences.
The nine lineages recovered here for T. albomarginatus s.l. correspond to eight geographic populations. Sympatric lineages D1 and D2, both indistinguishable morphologically, represent one geographical population. Each of the remaining seven lineages forms its own geographical one. These eight allopatric populations are distinguished by the combination of two meristic counts and three morphometric measurements (Figures 5, 6) and so represent eight operational taxonomic units (OTUs). Congruence in molecular and morphological evidence, coupled with concordant geographical distributions, supports the delineation of the nine lineages as eight putative species. It is evident that the currently recognized widespread species T. albomarginatus s.l. represents a species group including multiple species.
There is growing evidence for the existence of underestimated diversity when morphology-based species are scrutinized under a molecular phylogenetic context (Mongkolsamrit et al., 2020). Two or more distinct species are regarded to be cryptic if they are erroneously classified under one species name (Bickford et al., 2007). In most cases, cryptic species are those species that represent distinct lineages under molecular scrutiny but have been classified as a single species owing to failure to detect their subtle morphological differences (Jirsová et al., 2019). However, species of this kind should be referred to as pseudocryptic species (Matsuda and Gosliner, 2018) and cryptic species are truly represented by those species recently diverged, sympatrically occurring, and distinguishable only by DNA data (Brickford et al., 2007; Schnurr et al., 2018). Cases like this are also frequently seen in previous studies on freshwater fishes, but relatively common on invertebrates (Matsuda and Gosliner, 2018). Both cryptic and pseudocryptic species are herein unveiled within the T. albomarginatus species group. The species represented by the paired lineages (D1 and D2) are truly cryptic species, as both exhibit a distinct sequence divergence of 2.2% for the Cyt. b gene, a complete overlap in their ranges, and no external morphological differences. External morphology and allopatric distribution patterns distinguish the species represented by Lineages A2-A4 from the rest of this species group, and no previous studies have ever described them after a review of literature, thus making them pseudocryptic species.
Speciation and Rapid Diversification
The T. albomarginatus species group forms a monophyletic assemblage under rapid diversification (Figure 2). The majority of genetic divergences are found within five clades (A-E), but with only a small fraction of the genetic variation distributed among them, which is generally considered as evidence for rapid diversification (Groman, 2010; Sasaki et al., 2010). Clade A also shows a lower resolution of phylogenetic relationships among the four constituent lineages; the phylogeny displays a reticulate cladogenesis in the haplotype network (Figure 3), thereby indicating a phenomenon of rapid radiation (Cornejo and Scheidegger, 2015). Despite the lower genetic divergences here detected among these four lineages, they are distinguishable from morphology (Figures 5, 6) and represent four putative species.
The divergence time estimation among lineages A1 to A4 ranges from 1.35 to 1.72 Ma (Figure 3B), which coincides with the uprising of the Wuyi Mountains that likely occurred during the mid-Pleistocene (Zhang et al., 1990; Chiang et al., 2013). The rapid diversification within Clade A can best be interpreted as ecological adaptations of its included species to running waters as a result of mountain uprising. Our morphological examination indicates that the number of vertebrae is the most variable trait in the meristic counts (Figure 5). More vertebrae counts are found in Lineage A1 than Lineages A2-A4 (Figure 4). Consistent with this is the elevated distribution of the former when compared with the latter. It is likely that the increase in the vertebrae count of the species represented by Lineage A1 is an adaption to running water (such as flow velocity) in the Min-Jiang basin. It has been revealed that the number of vertebrae is correlated with ecological adaptations in freshwater fishes (Kimura et al., 2012; Kiso et al., 2012). Morphometric measurements such as head depth, outer mandibular-barbel length, and body depth at anus, which exhibit marked differences in the T. albomarginatus species group (Figures 5, 6), have been found to be related to ecological adaptations (Slaughter and Jacobson, 2008; Zeng and Liu, 2011; Pledger et al., 2014).
Three clades (A, B, and C), inhabiting in the lower Chang-Jiang basin and coastal rivers of Southeast China, are phylogenetically allied to each other but exhibit low sequence divergence and low resolution of phylogenetic relationship, thus indicating an explosive speciation event. The emergence of the most recent common ancestor of these three clades was estimated to be about 3.91 Ma, and the divergence time between Clades B and C was roughly 3.45 Ma. This period is broadly consistent with the intense uplifting of the Qinghai–Tibetan Plateau and the accompanying occurrence of Asian monsoons during the mid-Pliocene (3.6–2.6 Ma; Zhisheng et al., 2001; Zheng et al., 2002). Tectonic movement and climate change, which reshaped the geographic configuration of current drainage basins, are hypothesized to be key driving forces of vicariant events leading to the allopatric distribution of these clades. The geographic separation of the Poyang Lake system from streams on the southern bank of the lower Chang-Jiang in Anhui and Jiangsu provinces, triggered by the uplift of the Huangshan and Tianmu Mountains, is likely responsible for the disjunct distribution of Clades B and C, and from coastal rivers basin of Fujian Province, brought about by the rising of Wuyi-Yuhuan Mountains for that of Clades A and C. The disjunct distribution between Clades A and B is attributed to the formation of the watershed between streams in Zhejiang Province and Fujian Province (Figure 1). The effects of river capture on the expansion and dispersal of fishes were greatly reduced with the declining East Asian monsoon and lowered precipitation during the mid-Pliocene (Sun and Wang, 2005). Similar scenarios of regional divergence in South China during this timescale have previously been studied in some fish species, e.g., Hemiculter leucisculus (Chen et al., 2017) and Rhynchocypris oxycephalus (Yu et al., 2014).
Allopatry is also observed in Clade E and the remaining clades as well as Clades D and A + B + C. Clade E, occupying the Liu-Jiang of the middle Zhu-Jiang basin, is the earliest lineage to diverge in the T. albomarginatus species group. The time that Clade E separated from the rest of this species group was estimated to be around 4.92 Ma. Evidently, the disjunct distribution between this clade and others results from the formation of a water division between the middle Zhu-Jiang and middle Chang-Jiang basins and particularly between Liu-Jiang and Xiang-Jiang; the vicariant event responsible for this allopatric pattern was triggered by the upsurge of the Miaoling Mountains during the Early Pliocene. The disjunct distribution of the paired clades D and A + B + C can also be plausibly explained by geographical isolation; namely, the formation of the watershed between the Dongting and the Poyang Lake resulted from the uprising of Luoxiao-Mufu Mountains. Sympatric Lineages D1 and D2 have probably experienced allopatric divergence and subsequent merging. Continuous interbreeding between the two populations would lead them to have no autapomorphy to be diagnosed, just as suggested by Mishler et al. (2000).
Clade B has the most widespread distribution across many river basins, with its most northern extent at the Dongping Lake in Shandong Province (belonging to the Huang-He basin). It exhibits a similarity of genetic structure (Figures 2, 3), therefore indicating ongoing gene flow and dispersal among these hydrographic basins. This was made possible via the Beijing-Hangzhou Grand Canal, which was constructed more than 2,500 years ago (Chen et al., 2010). The canal starts from the lower Qiantang-Jiang of Zhejiang Province, across the lower Chang-Jiang, lower Huai-He, and lower Huang-He, to the Hai-He. The creation of the Beijing-Hangzhou Grand Canal makes it possible to have a northward-extended distribution as it has presently, given the presence of its closely allied lineages in South China: A in coastal rivers of Fujian Province and C in the Poyang Lake system.
Taxonomic Considerations
In this study, integrative taxonomy supports the detection of T. albomarginatus s.l. as including eight putative species. A conservative stance is adopted to recognize five taxa including four species, namely, T. albomarginatus (Clade B), T. analis (Clade C), T. lani (Clade E), and T. zhangfei, sp. nov. (Clade D), and a species complex (Clade A): the T. similis complex. Taxonomic confusions surrounding T. albomarginatus have been clarified by Cheng et al. (2021) based on their examination on the syntype and topotypes. The main characters typical for this species are shared with specimens from the lower reaches of the Chang-Jiang, Huai-He, and Qiantang-Jiang basins, and also coastal river basins of South Zhejiang Province. Samples from these rivers nested into Clade B in combined phylogenetic trees (Figure 2). Therefore, we have little doubt that Clade B represents the true T. albomarginatus.
The pairwise divergences of both the Cyt. b gene among Clades A-E range from 1.9 to 2.4%, which fulfill the barcode threshold of 2% (Ward, 2009) and are comparable to the pairwise divergence of 1.7% between T. aurantiacus and T. ondon (Ku et al., 2007). The substitution rate of the East Asian bagrid mitochondrial Cyt. b gene is estimated to vary from 0.18 to 0.30% sequence divergence per million years (Peng et al., 2002), much lower than that of other freshwater fishes, for example, 1.5% in salmonids (Shedlock et al., 1992), 0.68–1.0% for Cobitis species (Perdices and Doadrio 2001; Doadrio and Perdices 2005), 1.7% for Candidia barbatus (Wang et al., 2011), and 0.52–0.82% for cyprinid species (Zardoya and Doadrio, 1999; Ruber et al., 2007). This can explain, to a large extent, the relatively low sequence divergences here detected among five clades within the T. albomarginatus group when compared with non-bagrid species. The most recent common ancestor of T. albomarginatus group dates back to the early Pliocene when the complex paleogeographical history of East Asia, e.g., tectonic movement and climate change, facilitated the speciation of freshwater fishes. Thus, five main clades underwent recent rapid diversification, and so did the paired congeneric species T. pratti and T. truncatus (Figure 3B). Taking into account that Clade A was more recently diverged when compared with all other clades (in the middle Pleistocene), we follow the congruence of the time-calibrated frame to temporarily consider the A1-A4 lineages as a species complex.
Although the T. albomarginatus species group usually inhabits running waters of rivers, it has a limited swimming ability as indicated in their emarginated or round caudal fin instead of a deeply forked one in good swimmers such as T. fulvidraco which is widespread in mainstreams of East Asian rivers. The five taxa recognized here for this group are less widely distributed in tributaries of both the Chang-Jiang and Zhu-Jiang, but not found in their mainstreams; they also do not occur in the headwaters of the rivers they inhabit. Limited swimming ability and no occurrence in headwaters of nearby rivers indicate that inter-basin dispersal is highly unlikely, which leads to restricted gene flow between the clades. Diagnosable morphological differences detected in this study reinforce our confidence in the recognition of T. albomarginatus s.l. as containing multiple species.
The available name for the species represented by Clade C is Tachysurus analis, originally described by Nichols (1930) based on a single specimen of 101-mm SL collected from Hokou, northeastern Kiangsi (now Hekou Town, Yanshan County, Jiangxi Province), in an affluent of Lake Poyang-the Xin Jiang. Despite the current identification of T. analis as a valid species (Chu et al., 1999), it is known merely by the type specimen. Seemingly, no additional specimens have been reported from the Xin Jiang or elsewhere until now. Our photograph examination on the type (AMNH 6421) confirmed that T. analis, as stated in its original description, has a narrow, slightly tapering, rounded caudal fin (Figure 7A), a character not shared with all other congeneric species. In the molecular phylogenetic trees (Figure 2), samples caught from the Xin-Jiang (type locality of T. analis), Fu-He, and Gan-Jiang of the Poyang Lake system clustered into Clade C which was distantly allied with Clade B, representing a species distinct from T. albomarginatus. These samples also have a narrow, slightly tapering, rounded caudal fin (Figure 7B), a character uniquely diagnostic for T. analis; therefore, we consider them conspecific with T. analis. The most likely explanation for no reports of T. analis collected from the Xin-Jiang or the Poyang Lake system since its original description is misidentification as T. albomarginatus due to the shared presence of a broad white mark along the distal caudal-fin margin.

FIGURE 7. Lateral view of T. analis for: (A) AMNH 6421, hopotype; 101 mm SL; Hekou, Yanshan, Jiangxi Province, and (B) IHB201707014311, 124.2 mm SL, Yudu county, Jiangxi Province, in Gan-Jiang.
Recently, Cheng et al. (2021) confirmed that specimens previously misidentified as Tachysurus albomarginatus from the Liu-Jiang of the Zhu-Jiang basin belong to a new species named as T. lani. Samples from this river were recovered in this study to unite into Clade E. It is evident that T. lani is the available name for the species represented by this clade. Clade D, including two paired lineages B1 and B2, deserves specific status, given the recognition of their close genetic cluster (Clade E) as a valid species and also marked differences in the counts of vertebrae and anal-fin rays (Figure 5) with all other clades. A new species, T. zhangfei sp. nov., is here described for Clade D (see below). In reality, the new species represents a cryptic species complex, within which two cryptic species having overlapping distribution and indistinguishable morphology are included (Lineages D1 and D2).
Tachysurus similis, initially described by Nichols (1926) based on one specimen of 119 mm SL from the Min-Jiang, Yenping (now Nanping City in Fujian province) (Figure 8A), is the available name for the species represented by Lineage A1. This species has not been mentioned by subsequent researchers except Ferraris (2007), who listed it as species inquirenda in the Bagridae. The type (AMNH 8444) has 21 anal-fin rays rather than 17 as stated in the original description, and 47 vertebrae based on our observation on its X-ray photograph (Figure 8B). Our ongoing taxonomy studies of the currently recognized Chinese Tachysurus also show that T. analis is the only species with 47–49 vertebrae. Samples from Min-Jiang, with 19–22 anal-fin rays (Figure 5B) and 47-49 vertebrae (Figure 5A), nested into Lineage A1 and are distantly allied to Clades B and C (corresponding to T. albomarginatus and T. analis, respectively) in the phylogenetic trees (Figure 2). This clearly indicates that Lineage A1 represents a species distinct from either T. analis or T. albomarginatus. Therefore, the identity of specimens from the Min-Jiang previously misidentified as T. albomarginatus should actually be T. similis. Its validity has until now gone unrecognized, mainly owing to the imprecise original description, particularly in the count of anal-fin rays and the caudal-fin shape alike. The accompanied illustration (Page 1: Figure 1) depicts a species with a weakly forked instead of a slightly emarginate caudal fin (Figure 8C). To our knowledge, no specimens of Tachysurus have been found to possess 21 anal-fin rays and a weakly forked caudal fin. As such, the original description is likely erroneous.

FIGURE 8. Tachysurus similis: AMNH 8444, holotype, 119 mm SL; Nanping, Fujian Province, in lateral view (A) and X-ray (B) of body. IHB201809021142, topotype, mm SL, 152 mm SL Nanping, Fujian province (C).
As elaborated above, the three lineages (A2-A4), clustered within Clade A, represent three pseudocryptic species. However, it is premature to assign a specific status to each of these lineages, given that their sample sizes and number of sampling localities in this study are relatively small, especially for Lineages A2 and A3 (Figure 1). The names T. aff. similis Jin-Jiang, T. aff. similis Han-Jiang, and T. aff. similis Jiulong-Jiang are provisionally used to refer to the pseudocryptic species within the T. similis species complex.
Biodiversity Conservation
The detection of unrecognized diversity of Tachysurus albomarginatus s.l. in this analysis has many implications for biodiversity conservation. The species, as conventionally defined, has so far been listed as LC (least concern) in the recent assessment of Chinese freshwater fish red list (Zhang & Cao, 2021), owing to its wide distribution. It, however, has been demonstrated here to be a species group that can be further split into four species and a species complex, each with comparatively narrower distributions. This restricted distribution, one of the main reasons for concern in maintaining unique species in biodiversity conservation (Barlow et al., 2010), justifies concern on the long-term viability of these species or species complex. Tachysurus lani, represented by Clade E from the Liu-Jiang, has merely been collected in two sampling locations (locality 33 and 34, geographically separated by more than 200 km; see Figure 1) during our field surveys. The population of the T. aff. similis Jin-Jiang, represented by Lineage A2, is found only in one sampling location, with fewer than one dozen specimens caught during our field surveys into this river from 2018 to 2020. Tachysurus lani and the T. aff. similis Jin-Jiang, just like other rheophilic and predatory fishes, can be put under severe threat from anthropogenic disturbances such as river damming and illegal fishing. Thus, more efforts should be dedicated to protecting them, given that the single or fragmented habitats and small population sizes can result in lowered evolutionary potential and elevated extinction risk in the wild due to low genetic diversity (Cooper and Walters, 2010).
The revelation of hidden diversity for Tachysurus albomarginatus s.l. has ramifications not only for the protection of the detected putative species but also for regional biodiversity conservation of South China. Now, conservation efforts have been misdirected by the current misperception of freshwater fish diversity in the lowlands of South China where it is considered to have a lower proportion of endemic fish species. However, in this study, the currently recognized T. albomarginatus s.l. has been shown to exhibit a high level of geographic divergence. This highlights the need not only for molecular scrutiny of widely distributed species in lowland South China, particularly river-flooding plains, but also for adjustments of biodiversity conservation strategies to protect this largely overlooked fish diversity. Globally, freshwater ecosystem protection is subject to a territorial ecosystem scheme that puts emphasis on forests and land vertebrates; only a small portion of freshwater ecosystems is under protection (Hazen and Harris, 2007; Crous, 2013). South China is not the exception; in the framework of priority areas of territorial biodiversity conservation of China (Ma et al., 2010), merely the Poyang and Dongting lakes and headwaters of some rivers are included, and a small portion of freshwater fishes are thus protected. It is in urgent need to reconsider the biodiversity conservation plan of South China, particularly for freshwater ecosystems that are totally overlooked or ignored.
It is worthy of pointing out that the Tachysurus similis complex has the higher level of regional endemism in Fujian Province where each of the four major coastal river basins has its own putative species. This regional endemism is more or less repeated in many fish species such as Onychostoma barbatulum (Jang-Liaw and Chen, 2013), Acheilognathus macropterus (Yang et al., 2011), Sinibrama macrops (Zhao et al., 2013), and Squalidus argentatus (Yang et al., 2013). This evidently indicates that the coastal river basins of Fujian Province are a potential biodiversity hotspot in China and the members of T. similis complex should be considered as separate conservation units. In conclusion, it is apparent that priority should be given to the freshwater fishes of the coastal river basins of South China in biodiversity conservation plans.
Taxonomic Accounts
Tachysurus zhangfei sp. nov.
(Figure 9; Supplementary Figure S3, and Table 1).

FIGURE 9. (A) Lateral (B) dorsal and (C) ventral view of T. zhangfei in freshly captured specimen of 128.0 mm SL, from Tongren city, Guizhou province.
Leiocassis albomarginatus (not Rendahl, 1928): Ze, 1989: 249 (Jing-Jiang flowing into Yuan-Jiang of middle Chang-Jiang basin in Jiangkou County).
Holotype
IHB201709011724, 138.2 mm SL; South China: Guizhou Province: a stream tributary to Yuan-Jiang of middle Chang-Jiang basin in Songtao County.
Paratypes
IHB201709011717-23, 7 specimens, 76.6–154.2 mm SL; other data same as holotype.
Diagnosis
A member of the species group of Tachysurus defined by having a smooth anterior margin of the pectoral spine, short maxillary barbels not extending to the base of pectoral fin (Clade I of Ku et al., 2007), and an emarginate, truncate, or rounded caudal fin (Cheng et al., 2021: 12, Figure 10), with a broad white mark along the distal edge (Figures 7C, 8B, 9A). Tachysurus zhangfei is distinguished from all other species of this group by having fewer vertebrae (39–43 vs. 44–46 in T. albomarginatus and T. lani, or 47–49 in T. analis and T. similis; see Figure 5A), fewer anal-fin rays (15–18 vs. 19–21 in T. lani and the T. similis complex except for T. aff. similis Jin-Jiang or 20-23 in T. analis; see Figures 5A,B deeper body at anus (15.0–19.7 vs. 10.8–15.5% of SL in the T. similis complex; see Supplementary Figure S2A), a shallower head (depth 58.4–73.5 vs. 82.0–89.1% in T. albomarginatus; see Supplementary Figure S2B), and longer outer mandibular barbels (length 24.4–36.8 vs. 21.1–23.9% of HL in T. aff. similis Jin-Jiang, see Figure 6B).
Description
Morphometric data in Supplementary Table S1. See Figure 9 and Supplementary Figure S3 for general body appearance. Dorsal profile rising gradually, but not steeply from snout tip to dorsal-fin origin, then sloping evenly from there to posterior end of adipose-fin base and increasing ventrally to dorsal origin of procurrent caudal-fin rays. Ventral surface of head flattened, ventral profile of body straight or slightly convex from pectoral-fin insertion to anal-fin origin, decreasing evenly from posterior end of anal-fin base to origin of ventral procurrent caudal fin rays. Lateral line complete, straight, mid-lateral in position. Vetebrae 5 + 38–43.
Head depressed, broad, and covered with thin skin. Snout blunt in dorsal view, longer than eye diameter. Eye large, elliptical, covered with thick membrane, antero-lateral in head, visible when viewed dorsally, but not ventrally. Mouth inferior, crescentic; upper jaw projecting forward past lower jaw. Teeth villiform, in irregular rows on all tooth-bearing surfaces. Premaxillary tooth plates broad, of equal width throughout. Dentary tooth plates arched, broadest at symphysis, narrowing laterally, of same width at symphysis as premaxillary tooth plate. Vomerine tooth plate unpaired, continuous across midline, slightly curved anteriorly, much narrower than premaxillary tooth plate. Gill opening wide, extending from posttemporal to beyond isthmus. Barbels in four pairs; nasal ones small, thread-like, extending slightly beyond midpoint of eye, and maxillary ones slender, extending beyond posterior margin of eye, almost to gill membrane. Mandibular barbels in two pairs, thick, short; inner barbels longer than eye diameter. positioned in transverse row at level of posterior naris, not extending beyond anterior margin of eye; outer barbels rooted posterolateral to inner mandibular barbell, extending beyond posterior margin of eye.
Dorsal fin with spinelet, spine, and seven soft, branched rays, inserted nearer to anal-fin origin than to snout tip or nearer to pectoral-fin insertion than to pelvic-fin insertion, closer to anal-fin origin than to snout tip. Spinelet flattened, with long, blunt distal tip. Dorsal-fin spine stout, with smooth anterior margin and slightly serrated posterior margin distally, longer than pectoral-fin spine. First dorsal-fin soft ray longest, surpassing tip of last ray. Distal margin of dorsal fin rays nearly straight. Nuchal plate triangular, with a pointed tip. Adipose fin inserted behind vertical through anal-fin origin, with convex distal margin for its entire length and a deeply incised posterior part to form a rounded apex. Adipose-fin base moderately long, longer than anal-fin base length.
Pectoral fin with a spine and eight soft branched rays, inserted anterior to vertical through posteriormost point of opercle, extending for half of distance to base of pelvic-fin spine. Pectoral-fin spine stout, sharply pointed at tip, shorter than dorsal-fin spine, with a smooth anterior margin and 12-15 (mean 13.3) strong serrations along posterior margin. Pectoral-fin margin straight anteriorly, convex posteriorly. Cleithral process triangular with a sharp pointed tip. Pelvic fin with five soft branched rays, inserted equidistant from vertical through caudal-fin base and tip of snout. Tip of depressed pelvic fin slightly extending beyond anal-fin origin. Pelvic-fin distal margin convex. Anus and urogenital opening halfway between anal-fin origin and pelvic-fin insertion.
Anal fin shorter than adipose-fin base in length, with 15 (4), 16 (12), 17 (9), or 18 (5) branched rays; inserted anterior to adipose-fin origin. Anal-fin origin nearer to caudal-fin base than to tip of snout. Distal margin of anal fin convex; anterior rays shortest.
Caudal fin broadly rounded or slightly emarginate, with 8 + 9 principal rays; upper lobe roughly equidistant from lower one; procurrent rays extending slightly anterior to fin base.
Coloration
In formalin-preserved specimens (Figure 9), body dark brown dorsally and laterally, grayish ventrally; caudal and anal fins blackish with a broad white distal margin, and other fins uniformly dark brown. When in life (Supplementary Figure S3A), body pale brown dorsally and laterally, yellowish ventrally; caudal and anal fins with a broad yellowish or white distal margin, and other fins uniformly brown.
Distribution
Tachysurus zhangfei is currently known from the Xiang-Jiang and Yuan-Jiang, and possibly Dongting Lake, in the middle Chang-Jiang basin.
Etymology
The specific epithet, used as a noun, is named after Zhang Fei, a character of the Romance of the Three Kingdoms—one of four famous literature works in China, in allusion to the presence of a blackish or brown body. He is endowed with an imagination of a black face in China opera.
Taxonomic Notes
The taxonomic usefulness of the presence or absence of serrations along the anterior margin of the pectoral spine and length of maxillary barbels was suggested in Ku et al., 2007 phylogenetic analysis of Pseudobagrus (=Tachysurus) predicated on mitochondrial sequence data. Clade I (Page, 156: Figure 1) recovered by them was formed by species with a smooth anterior margin of the pectoral spine and short maxillary barbels not extending to the base of the pectoral spine. Twenty-one constituent species can be subdivided into two groups based on the caudal-fin shape (Cheng et al., 2021). The species group with an emarginate, truncate, or rounded caudal fin, to which eight putative species recovered here for Tachysurus albomarginatus are referred, includes the following nine species: T. adiposalis, T. brachyrhabdion, T. gracilis, T. omeihensis, T. tenuis, T. trilineatus, T. truncatus, and T. ussuriensis. All these species except T. adiposalis and T. omeihensis were included in the ingroup of our molecular phylogenetic analysis. The topology of the phylogenetic tree (Figure 2) shows the monophyletic nature of the T. albomarginatus group. It is apparent that the caudal-fin coloration can be used as a character uniquely diagnostic for this species group. A narrow white mark along the distal margin of the caudal fin is present in T. tenius and T. ussuriensis (Cheng et al., 2021). A short narrow white mark is seen along the upper/lower margin of the caudal fin in both T. brachyrhabdion and T. gracilis (Li et al., 2005; Cheng et al., 2008).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Ethics Committee of Institute of Hydrabiology, Chinese academy of sciences.
Author Contributions
W-HS and J-LC measured the specimens, analyzed the data, and prepared the manuscript. EZ contributed to the funding of research and organization of the manuscript. All authors read and approved the final version of the manuscript.
Funding
This study was funded by two special foundations for Biodiversity Survey, Monitoring and Assessment (2017HB2096001006 and 2019HB2096001006), and one special foundation for Natural Science and Technology Basic Research Program of China (2019FY10904).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are grateful to Y. Li for collecting specimens. We thank C. Huang for English corrections and suggestions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.713793/full#supplementary-material
Supplementary Figure 1 | Principal components analysis (PCA) scores of two sympatric lineages belonging to Clade D.
Supplementary Figure 2 | Relation between (A) body depth at anus and SL for T. zhangfei and the T. similis complex; relation between (B) Head depth and HL for T. zhangfei and T. albomarginatus.
Supplementary Figure 3 | Lateral (A), dorsal (B) and (C) ventral view of Tachysurus nicholsi sp. nov., IHB 201709011724, holotype, 138.2 mm SL.
References
Akaike, H. T. (1974). A New Look at the Statistical Model Identification. Automat. Cont. IEEE Trans. 19 (6), 716–723. doi:10.1007/978-1-4612-1694-0_16
Barlow, J., Gardner, T. A., Louzada, J., and Peres, C. A. (2010). Measuring the Conservation Value of Tropical Primary Forests: The Effect of Occasional Species on Estimates of Biodiversity Uniqueness. PLoS ONE 5 (3), e9609. doi:10.1371/journal.pone.0009609
Bickford, D., Lohman, D. J., Sodhi, N. S., Ng, P. K. L., Meier, R., Winker, K., et al. (2007). Cryptic Species as a Window on Diversity and Conservation. Trends Ecol. Evol. 22, 148–155. doi:10.1016/j.tree.2006.11.004
Chen, H. X. (1984). The Fishes of Fujian Province, China. Fuzhou: Fujian Science and Technology Publishing House press. (in Chinese).
Chen, W., Zhong, Z., Dai, W., Fan, Q., He, S., and Chen, W. (2017). Phylogeographic Structure, Cryptic Speciation and Demographic History of the Sharpbelly (Hemiculter leucisculus), a Freshwater Habitat Generalist from Southern China. BMC Evol. Biol. 17 (1), 216. doi:10.1186/s12862-017-1058-0
Chen, X. L., Chiang, T. Y., Lin, H. D., Zheng, H. S., Shao, K. T., and Zhang, Q. (2010). Mitochondrial DNA Phylogeography of Glyptothorax Fokiensis and Glyptothorax Hainanensis in Asia. J. Fish Biol. 70, 75–93.
Cheng, J.-L., Ishihara, H., and Zhang, E. (2008). Pseudobagrus Brachyrhabdion, a New Catfish (Teleostei: Bagridae) from the Middle Yangtze River Drainage, South China. Ichthyol. Res. 55 (2), 112–123. doi:10.1007/s10228-007-0020-3
Cheng, J. L., Shao, W. H., Anders, Lopez. J., and Zhang, E. (2021). Tachysurus lani, a New Catfish Species (Teleostei: Bagridae) from the Pearl River basin, South China. Ichthylogical Exploration of Freshwaters 1156, 1–17. doi:10.23788/ief-1156
Cheng, Q. T., and Zhou, C. W. (1997). The Fishes of Shandong Province. Ji’nan: Shandong Science and Technology Publishing House press. (in Chinese).
Chu, X. L., Zheng, B. S., and Dai, D. Y. (1999). FAUNA SINICA Osteichthys Siluriformes. Beijing: Science press. (in Chinese).
Chiang, T.-Y., Lin, H.-D., Zhao, J., Kuo, P.-H., Lee, T.-W., and Hsu, K.-C. (2013). Diverse Processes Shape Deep Phylogeographical Divergence in Cobitis sinensis (Teleostei: Cobitidae) in East Asia. J. Zoolog. Syst. Evol. Res. 51, 316–326. doi:10.1111/jzs.12030
Clement, M., Posada, D., and Crandall, K. A. (2000). TCS: a Computer Program to Estimate Gene Genealogies. Mol. Ecol. 9 (10), 1657–1659. doi:10.1046/j.1365-294x.2000.01020.x
Cooper, J., and Walters, M. (2010). Iterated Point-Line Configurations Grow Doubly-Exponentially. Discrete Comput. Geom 43 (3), 554–562. doi:10.1007/s00454-009-9208-9
Cornejo, C., and Scheidegger, C. (2015). Multi-gene Phylogeny of the genusLobaria: Evidence of Species-Pair and Allopatric Cryptic Speciation in East Asia. Am. J. Bot. 102 (12), 2058–2073. doi:10.3732/ajb.1500207
Crous, C. J. (2013). The Mesofilter Concept and Biodiversity Conservation in Afro-Montane Grasslands. Stellenbosch Stellenbosch Univ. 17 (June), 165–181.
da Silva, R., Peloso, P. L. V., Sturaro, M. J., Veneza, I., Sampaio, I., Schneider, H., et al. (2018). Comparative Analyses of Species Delimitation Methods with Molecular Data in Snappers (Perciformes: Lutjaninae). Mitochondrial DNA A 29 (7), 1108–1114. doi:10.1080/24701394.2017.1413364
Dao, W., Li, X., Yang, H. F., and Zhou, W. (2020). Pareuchiloglanis (Teleostei: Sisoridae) from the Pearl River, China with Description of Three New Species. J. Fish. Biol. 96 (1), 23–36. doi:10.1111/jfb.14178
Darwall, W., Bremerich, V., De Wever, A., Dell, A. I., Freyhof, J., Gessner, M. O., et al. (2018). TheAlliance for Freshwater Life: A Global Call to Unite Efforts for Freshwater Biodiversity Science and Conservation. Aquat. Conserv: Mar Freshw Ecosyst 28 (4), 1015–1022. doi:10.1002/aqc.2958
Das, I. (2007). Cryptic Species as a Window on Diversity and Conservation. Trends Ecol. Evol. 22 (3), 148–155.
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). Jmodeltest 2: More Models, New Heuristics and Parallel Computing. Nature Methods 9 (8), 772. doi:10.1038/nmeth.2109
Dawar, F. U., Babu V, S., Kou, H., Qin, Z., Wan, Q., Zhao, L., et al. (2019). The RAG2 Gene of Yellow Catfish (Tachysurus fulvidraco) and its Immune Response against Edwardsiella Ictaluri Infection. Develop. Comp. Immunol. 98, 65–75. doi:10.1016/j.dci.2019.04.006
Delic, T., Trontelj, P., Rendoš, M., and Fišher, C. (2017). The Importance of Naming Cryptic Species and the Conservation of Endemic Subterranean Amphipods. Scientific Rep. 7 (1), 3391. doi:10.1038/s41598-017-02938-z
Doadrio, I., and Perdices, A. (2005). Phylogenetic Relationships Among the Ibero-African Cobitids (Cobitis, Cobitidae) Based on Cytochrome B Sequence Data. Mol. Phylogenet. Evol. 37, 484–493. doi:10.1016/j.ympev.2005.07.009
Drummond, A. J., and Rambaut, A. (2007). BEAST: Bayesian Evolutionary Analysis by Sampling Trees. BMC Evol. Biol. 7 (1), 214. doi:10.1186/1471-2148-7-214
Du, L. N., Yang, J., Min, R., Chen, X. Y., and Yang, J. X. (2021). A Review of the Cypriniform Tribe Yunnanilini Prokofiev, 2010 from China, with an Emphasis on Five Genera Based on Morphologies and Complete Mitochondrial Genomes of Some Species. Zool Res. 42 (3), 310–334. doi:10.24272/j.issn.2095-8137.2020.229
Edgar, R. C. (2004). MUSCLE: a Multiple Sequence Alignment Method with Reduced Time and Space Complexity. Bmc Bioinformatics 5 (1), 113. doi:10.1186/1471-2105-5-113
Eme, D., Zagmajster, M., Delić, T., Fišer, C., Flot, J.-F., Konecny-Dupré, L., et al. (2017). Do cryptic Species Matter in Macroecology? Sequencing European Groundwater Crustaceans Yields Smaller Ranges but Does Not challenge Biodiversity Determinants. Ecography 41, 424–436. doi:10.1111/ecog.02683
Ferraris, Carl. J. (2007). Checklist of Catfishes, Recent and Fossil (Osteichthyes: Siluriformes), and Catalogue of Siluriform Primary Types. Zootaxa 1418, 81–107. doi:10.11646/zootaxa.1418.1.1
Forister, M. L., Nice, C. C., Fordyce, J. A., Gompert, Z., and Shapiro, A. M. (2008). Considering Evolutionary Processes in the Use of Single-Locus Genetic Data for Conservation, with Examples from the Lepidoptera. J. Insect Conserv 12 (1), 37–51. doi:10.1007/s10841-006-9061-6
Gao, G. F. (1990). The Freshwater Fishes of Guangdong Province, China. Guangzhou: Guangdong Science and Technology Publishing House Press.
Gascón, S., Boix, D., Sala, J., and Quintana, X. D. (2008). Relation between Macroinvertebrate Life Strategies and Habitat Traits in Mediterranean Salt Marsh Ponds (Empordà Wetlands, NE Iberian Peninsula). Hydrobiologia 597 (1), 71–83. doi:10.1007/s10750-007-9215-x
Groman, Pellmyr. (2010). Rapid Evolution and Specialization Following Host Colonization in a Yucca Moth. J. Evol. Biol. 13 (2), 223–236. doi:10.1046/j1420-9101.2000.00159.x
Harrison, I. J., Green, P. A., Farrell, T. A., Juffe-Bignoli, D., Sáenz, L., and Vörösmarty, C. J. (2016). Protected Areas and Freshwater Provisioning: a Global Assessment of Freshwater Provision, Threats and Management Strategies to Support Human Water Security. Aquat. Conserv: Mar. Freshw. Ecosyst. 26, 103–120. doi:10.1002/aqc.2652
Hazen, H. D., and Harris, L. M. (2007). Limits of Territorially-Focused Conservation: a Critical Assessment Based on Cartographic and Geographic Approaches. Environ. Conservation 34 (4), 280–290. doi:10.1017/s0376892907004237
Jang-Liaw, N.-H., and Chen, I.-S. (2013). Onychostoma minnanensis, a New Cyprinid Species (Teleostei: Cyprinidae) from Fujian, Southern mainland China, with Comments on the Mitogenetic Differentiation Among Related Species. Ichthyol. Res. 60 (1), 62–74. doi:10.1007/s10228-012-0312-0
Jirsová, D., Štefka, J., Blažek, R., Malala, J. O., Lotuliakou, D. E., Mahmoud, Z. N., et al. (2019). From Taxonomic Deflation to Newly Detected Cryptic Species: Hidden Diversity in a Widespread African Squeaker Catfish. Sci. Rep. 9 (1), 15748. doi:10.1038/s41598-019-52306-2
Kapli, P., Lutteropp, S., Zhang, J., Kobert, K., Pavlidis, P., Stamatakis, A., et al. (2017). Multi-rate Poisson Tree Processes for Single-Locus Species Delimitation under Maximum Likelihood and Markov Chain Monte Carlo. Bioinformatics 33, 1630–1638. doi:10.1093/bioinformatics/btx025
Kekkonen, M., and Hebert, P. D. N. (2014). DNA Barcode‐based Delineation of Putative Species: Efficient Start for Taxonomic Workflows. Mol. Ecol. Resour. 14 (4), 706–715. doi:10.1111/1755-0998.12233
Kimura, M. (1980). A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 16, 111–120. doi:10.1007/bf01731581
Kimura, T., Shinya, M., and Naruse, K. (2012). Genetic Analysis of Vertebral Regionalization and Number in Medaka (Oryzias latipes) Inbred Lines. G3 Genesgenetics 2 (11), 1317–1323. doi:10.1534/g3.112.003236
Kiso, S., Miyake, T., and Yamahira, K. (2012). Heritability and Genetic Correlation of Abdominal and Caudal Vertebral Numbers in Latitudinal Populations of the Medaka Oryzias latipes. Environ. Biol. Fish. 93 (2), 185–192. doi:10.1007/s10641-011-9904-1
Ku, X., Peng, Z., Diogo, R., and He, S. (2007). MtDNA Phylogeny Provides Evidence of Generic Polyphyleticism for East Asian Bagrid Catfishes. Hydrobiologia 579 (1), 147–159. doi:10.1007/s10750-006-0401-z
Laisí, C. G., Tiago, C. P., Naiara, G. S., Paulo, S. P., and Daniel, C. C. (2015). Integrative Taxonomy Detects Cryptic and Overlooked Fish Species in a Neotropical River Basin. Genetica 143, 581–588. doi:10.1007/s10709-015-9856-z
Lei, Y., Mayden, R. L., and He, S. (2009). Population Genetic Structure and Geographical Differentiation of the Chinese Catfish Hemibagrus macropterus (Siluriformes, Bagridae): Evidence for Altered Drainage Patterns. Mol. Phylogenet. Evol. 51 (2), 405–411. doi:10.1016/j.ympev.2009.01.004
Lessios, H. A. (2008). The Great American Schism: Divergence of marine Organisms after the Rise of the Central American Isthmus. Annu. Rev. Ecol. Evol. Syst. 39 (1), 63–91. doi:10.1146/annurev.ecolsys.38.091206.095815
Li, J., Chen, X., and Chan, B. P. L. (2005). A New Species of Pseudobagrus (Teleostei: Siluriformes: Bagridae) from Southern China. Zootaxa 1067, 49–57. doi:10.11646/zootaxa.1067.1.3
Li, X., Dao, W., and Zhou, W. (2020). Type Locality and Species Identity of Pareuchiloglanis Sinensis (Hora & Silas) with a Description of a New Species of the Genus from the Upper Yangtze River basin in Southern China. J. Fish. Biol. 97 (3), 827–844. doi:10.1111/jfb.14438
Liang, H., Guo, S., Luo, X., Li, Z., and Zou, G. (2016). Molecular Diagnostic Markers of Tachysurus fulvidraco and Leiocassis Longirostris and Their Hybrids. SpringerPlus 5 (1), 2115. doi:10.1186/s40064-016-3766-0
Librado, P., and Rozas, J. (2009). DnaSP V5: a Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 25, 1451–1452. doi:10.1093/bioinformatics/btp187
Ma, Z., Li, B., Li, W., Han, N., and Watkinson, C. (2010). Conflicts between Biodiversity Conservation and Development in a Biosphere reserve. J. Appl. Ecol. 46 (3), 527–535. doi:10.1111/j.1365-2664.2008.01528.x
Matsuda, S. B., and Gosliner, T. M. (2018). Molecular Phylogeny of Glossodoris (Ehrenberg, 1831) Nudibranchs and Related Genera Reveals Cryptic and Pseudocryptic Species Complexes. Cladistics 34, 41–56. doi:10.1111/cla.12194
Mishler, B. D. (2000). Deep Phylogenetic Relationships Among “Plants” and their Implications for Classification. TAXON 49 (4). doi:10.2307/1223970
Mongkolsamrit, S., Noisripoom, W., Tasanathai, K., Khonsanit, A., Thanakitpipattana, D., Himaman, W., et al. (2020). Molecular Phylogeny and Morphology Reveal Cryptic Species in Blackwellomyces and Cordyceps (Cordycipitaceae) from Thailand. Mycol. Prog. 19 (9), 957–983. doi:10.1007/s11557-020-01615-2
Ng, H. H., and Freyhof, J. (2007). Pseudobagrus nubilosus, a New Species of Catfish from central Vietnam (Teleostei:Bagridae), with Notes on the Validity of Pelteobagrus and Pseudobagrus. Ichthyol. exploration Freshwaters 18 (1), 9–16. doi:10.1093/icesjms/fsl022
Ng, H. H., and Kottelat, M. (2007). The Identity of Tachysurus sinensis La Cepède, 1803, with the Designation of a Neotype (Teleostei: Bagridae) and Notes on the Identity of T. Fulvidraco (Richardson, 1845). Electron. J. Ichthyology 2, 35–45.
Nichols, J. T. (1926). Some Chinese Fresh-Water Fishes. XV. Two Apparently Undescribed Catfishes from Fukien. XVI. Concerning Gudgens Related to Pseudogobio, and Two New Species of it. XVII. Two New Rhodeins. Amur. Mus. Novitates, 1–7.
Ozkilinc, H., and Sevinc, U. (2018). Molecular Phylogenetic Species in Alternaria Pathogens Infecting Pistachio and Wild Relatives. 3 Biotech. 8 (5), 250. doi:10.1007/s13205-018-1277-0
Peng, Z., He, S., and Zhang, Y. (2002). Mitochondrial Cytochrome B Sequence Variations and Phylogeny of the East Asian Bagrid Catfishes. Prog. Nat. Sci. 12 (6), 421–425.
Perdices, A., and Doadrio, I. (2001). The Molecular Systematics and Biogeography of the European Cobitids Based on Mitochondrial DNA Sequences. Mol. Phylogenet. Evol. 19, 468–478. doi:10.1006/mpev.2000.0900
Perdices, A., Sayanda, D., and Coelho, M. M. (2005). Mitochondrial Diversity of Opsariichthys Bidens (Teleostei, Cyprinidae) in Three Chinese Drainages. Mol. Phylogenet. Evol. 37 (3), 920–927. doi:10.1016/j.ympev.2005.04.020
Pledger, A. G., Rice, S. P., and Millett, J. (2014). Reduced Bed Material Stability and Increased Bedload Transport Caused by Foraging Fish: a Flume Study with Juvenile Barbel (Barbus barbus). Earth Surf. Process. Landforms 39 (11), 1500–1513. doi:10.1002/esp.3592
Posada, D., and Crandall, K. A. (2001). Intraspecific Gene Genealogies: Trees Grafting into Networks. Trends Ecol. Evol. 16 (1), 37–45. doi:10.1016/s0169-5347(00)02026-7
Puillandre, N., Lambert, A., Brouillet, S., and Achaz, G. (2012). ABGD, Automatic Barcode Gap Discovery for Primary Species Delimitation. Mol. Ecol. 21, 1864–1877. doi:10.1111/j.1365-294x.2011.05239.x
Reist, J. D. (1985). An Empirical Evaluation of Several Univariate Methods that Adjust for Size Variation in Morphometric Data. Can. J. Zool. 63, 1429–1439. doi:10.1139/z85-213
Rendahl, H. (1928). Beiträge zur kenntnis der Chinesischen Süsswasserfische. I. Systematischer Teil. Arkiv för Zoologi 20, 1–194.
Rojas, P., Scott, S., Tobar, I., Romero, R., and Vila, I. (2020). Head Morphometry of Orestias (Cyprinodontiformes). Response to Extreme Southern Altiplano Systems? Environ. Biol. Fishes 103 (9), 953–964. doi:10.1007/s10641-020-00997-2
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian Phylogenetic Inference under Mixed Models. Bioinformatics 19 (12), 1572–1574. doi:10.1093/bioinformatics/btg180
Rüber, L., Kottelat, M., Tan, H., Ng, P. K., and Britz, R. (2007). Evolution of Miniaturization and the Phylogenetic Position of Paedocypris, Comprising the World's Smallest Vertebrate. BMC Evol. Biol. 7, 38. doi:10.1186/1471-2148-7-38
Sasaki, T., Tsubakishita, S., Tanaka, Y., Sakusabe, A., Ohtsuka, M., and Hirotaki, S. (2010). Multiplex-pcr Method for Species Identification of Coagulase-Positive Staphylococci. J. Clin. Microbiol. 48 (3), 765–769. doi:10.1128/JCM.01232-09
Schmidt, R. C., Dillon, M. N., Kuhn, N. M., Bart, H. L., and Pezold, F. (2019). Unrecognized and Imperilled Diversity in an Endemic Barb (Smiliogastrini, Enteromius ) from the Fouta Djallon highlands. Zool Scr 48 (5), 605–613. doi:10.1111/zsc.12362
Schnurr, S., Osborn, K. J., Malyutina, M., Jennings, R., Brix, S., Driskell, A., et al. (2018). Hidden Diversity in Two Species Complexes of Munnopsid Isopods (Crustacea) at the Transition between the Northernmost North Atlantic and the Nordic Seas. Mar. Biodiversity 48 (14), 813–843. doi:10.1007/s12526-018-0877-6
Shedlock, A. M., Parker, J. D., Crispin, D. A., Pietsch, T. W., and Burmer, G. C. (1992). Evolution of the Salmonid Mitochondrial Control Region. Mol. Phylogenet. Evol. 1, 179–192. doi:10.1016/1055-7903(92)90014-8
Silvestro, D., and Michalak, I. (2012). Raxmlgui: A Graphical Front-End for Raxml. Organisms Diversity & Evolution 12 (4), 335–337. doi:10.1007/s13127-011-0056-0
Slaughter, J. E., and Jacobson, B. (2008). Gape: Body Size Relationship of Flathead Catfish. North Am. J. Fish. Manag. 28 (1), 198–202. doi:10.1577/m06-033.1
Sun, X. J., and Wang, P. X. (2005). How Old Is the Asian Monsoon System? Palaeobotanical Records from China. Palaeogeogr. Palaeocl 222 (3), 181–222. doi:10.1016/j.palaeo.2005.03.005
Tan, D. S. H., Ang, Y., Lim, G. S., Ismail, M. R. B., and Meier, R. (2010). From €˜cryptic Species’ to Integrative Taxonomy: an Iterative Process Involving DNA Sequences, Morphology, and Behaviour Leads to the Resurrection ofSepsis pyrrhosoma(Sepsidae: Diptera). Zoologica Scripta 39 (1), 51–61. doi:10.1111/j.1463-6409.2009.00408.x
Wang, C.-F., Hsieh, C.-H., Lee, S.-C., and Wang, H.-Y. (2011). Systematics and Phylogeography of the Taiwanese Endemic Minnow Candidia barbatus (Pisces: Cyprinidae) Based on DNA Sequence, Allozymic, and Morphological Analyses. Zool J. Linn. Soc. 161, 613–632. doi:10.1111/j.1096-3642.2010.00646.x
Ward, R. D. (2009). DNA Barcode Divergence Among Species and Genera of Birds and Fishes. Mol. Ecol. Resour. 9 (4), 1077–1085. doi:10.1111/j.1755-0998.2009.02541.x
Welton, L. J., Travers, S. L., Siler, C. D., and Brown, R. M. (2014). Integrative Taxonomy and Phylogeny-Based Species Delimitation of Philippine Water Monitor Lizards (Varanus salvator Complex) with Descriptions of Two New Cryptic Species. Zootaxa 3881 (3), 201–227. doi:10.11646/zootaxa.3881.3.1
Xiao, W., Zhang, Y., and Liu, H. (2001). Molecular Systematics of Xenocyprinae (Teleostei: Cyprinidae): Taxonomy, Biogeography, and Coevolution of a Special Group Restricted in East Asia. Mol. Phylogenet. Evol. 18 (2), 163–173. doi:10.1006/mpev.2000.0879
Yang, J.-Q., Hsu, K.-C., Liu, Z.-Z., Su, L.-W., Kuo, P.-H., Tang, W.-Q., et al. (2016). The Population History of Garra Orientalis (Teleostei: Cyprinidae) Using Mitochondrial DNA and Microsatellite Data with Approximate Bayesian Computation. BMC Evol. Biol. 16 (1), 73. doi:10.1186/s12862-016-0645-9
Yang, J.-Q., Tang, W.-Q., Sun, Y., Tsai, K.-C., Zhou, Z.-C., Liu, Z.-Z., et al. (2013). Microsatellite Diversity and Population Genetic Structure of Squalidus argentatus (Cyprinidae) on the Island of Hainan and mainland China. Biochem. Syst. Ecol. 50, 7–15. doi:10.1016/j.bse.2013.03.023
Yang, Q., Zhu, Y., Xiong, B., and Liu, H. (2011). Acheilognathus Changtingensissp. nov., a New Species of the Cyprinid GenusAcheilognathus(Teleostei: Cyprinidae) from Southeastern China Based on Morphological and Molecular Evidence. Zoolog. Sci. 28 (2), 158–167. doi:10.2108/zsj.28.158
Yu, D., Chen, M., Tang, Q., Li, X., and Liu, H. (2014). Geological Events and Pliocene Climate Fluctuations Explain the Phylogeographical Pattern of the Cold Water Fish Rhynchocypris Oxycephalus (Cypriniformes: Cyprinidae) in China. BMC Evol. Biol. 14, 225. doi:10.1186/s12862-014-0225-9
Zardoya, R., and Doadrio, I. (1999). Molecular Evidence on the Evolutionary and Biogeographical Patterns of European Cyprinids. J. Mol. Evol. 49, 227–237. doi:10.1007/pl00006545
Zeng, Y., and Liu, H. (2011). The Evolution of Pharyngeal Bones and Teeth in Gobioninae Fishes (Teleostei: Cyprinidae) Analyzed with Phylogenetic Comparative Methods. Hydrobiologia 664 (1), 183–197. doi:10.1007/s10750-010-0598-8
Zhang, H. N., Chen, C. G., Huang, K. R., Li, Z. Q., Zhang, F. L., and Chen, G. Z. (1990). The New Geological Structures, Tectonic Movements and Geological Environment in Coastal Line of South China. Beijing, Earthquake Press. (in Chinese).
Zhang, E., and Cao, W. X. (2021). China's Red List of Biodiversity: Vertebrates, Volume V, Freshwater Fishes. Beijing: Science press. (in Chinese).
Zhao, L., Zhou, X., Liu, Q., and Zhang, H. (2013). Genetic Variation and Phylogeography of Sinibrama macrops (Teleostei: Cyprinidae) in Qiantang River Basin, China. Biochem. Syst. Ecol. 49, 10–20. doi:10.1016/j.bse.2013.01.007
Zheng, B., Xu, Q., and Shen, Y. (2002). The Relationship between Climate Change and Quaternary Glacial Cycles on the Qinghai-Tibetan Plateau: Review and Speculation. Quat. Int. 97-98, 93–101. doi:10.1016/s1040-6182(02)00054-x
Keywords: cryptic species, allopatric speciation, integrative taxonomy, species delineation, freshwater fishes
Citation: Shao W-H, Cheng J-L and Zhang E (2021) Eight in One: Hidden Diversity of the Bagrid Catfish Tachysurus albomarginatus s.l. (Rendhal, 1928) Widespread in Lowlands of South China. Front. Genet. 12:713793. doi: 10.3389/fgene.2021.713793
Received: 24 May 2021; Accepted: 19 October 2021;
Published: 17 November 2021.
Edited by:
Stephane Boissinot, New York University Abu Dhabi, United Arab EmiratesReviewed by:
Heok Hee Ng, National University of Singapore, SingaporeYann Bourgeois, University of Portsmouth, United Kingdom
Copyright © 2021 Shao, Cheng and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: E Zhang, zhange@ihb.ac.cn
 Wei-Han Shao
Wei-Han Shao Jian-Li Cheng
Jian-Li Cheng E Zhang
E Zhang