- 1Department of Agriculture and Animal Health, School of Agriculture and Life Sciences, College of Agriculture and Environmental Sciences, University of South Africa, Roodeport, South Africa
- 2Department of Botany, Nelson Mandela University, Port Elizabeth, South Africa
- 3Research Support Services, North-West Provincial Department of Agriculture and Rural Development, Potchefstroom, South Africa
- 4Department of Biochemistry, School of Mathematical and Natural Sciences, University of Venda, Thohoyandou, South Africa
The causal agent of rust, Uromyces appendiculatus is a major constraint for common bean (Phaseolus vulgaris) production. This pathogen causes substantial yield losses in many common bean production areas worldwide. U. appendiculatus is widely distributed and although there have been numerous breakthroughs in breeding for resistance, its ability to mutate and evolve still poses a major threat to common bean production. An understanding of plant phytochemical properties can aid in accelerating breeding for rust resistance. In this study, metabolome profiles of two common bean genotypes Teebus-RR-1 (resistant) and Golden Gate Wax (susceptible) were investigated for their response to U. appendiculatus races (1 and 3) at 14- and 21-days post-infection (dpi) using liquid chromatography-quadrupole time-of-flight tandem mass spectrometry (LC-qTOF-MS). Non-targeted data analysis revealed 71 known metabolites that were putatively annotated, and a total of 33 were statistically significant. Key metabolites including flavonoids, terpenoids, alkaloids and lipids were found to be incited by rust infections in both genotypes. Resistant genotype as compared to the susceptible genotype differentially enriched metabolites including aconifine, D-sucrose, galangin, rutarin and others as a defence mechanism against the rust pathogen. The results suggest that timely response to pathogen attack by signalling the production of specific metabolites can be used as a strategy to understand plant defence. This is the first study to illustrate the utilization of metabolomics to understand the interaction of common bean with rust.
1 Introduction
Common bean (Phaseolus vulgaris) is one of the most important human and animal consumable legume worldwide (Vega et al., 2017; Sadohara, 2020). Globally, common bean is cultivated on about 30 million hectares (Mha), while in Africa over 7.5 Mha are cultivated (Mukankusi et al., 2019). Common bean rust, a disease originating from the fungal pathogen Uromyces appendiculatus species affects the production of common bean in many production areas by causing between mild and severe damage to plants upon infection (Delgado et al., 2013). The urediniospores of U. appendiculatus can survive in winter and can be a source of inoculum in summer when the humidity and temperature are favourable (Gross and Venette, 2001; Mukankusi et al., 2019). The severity of the pathogen may occur in cooler environments with temperatures of about 17°C–20°C and high humidity (Liebenberg et al., 2005). It is difficult to predict yield losses from rust infestation, the loss of yields of up to 100% may occur under severe disease pressure (Lindgren et al., 1995; Singh and Schwartz, 2010). Gross and Venette (2001) also reported that even yield losses of about 6% can have reduced financial gain to farmers growing the crop on a larger scale. Currently, climate changes favour rust spreads in common bean producing areas as spores favourably flow in the air from one area to the other (Alleyne et al., 2008; Singh and Schwartz, 2010). Singh and Schwartz (2010) also added that plant debris left over from the previous season can also be a source of the disease. Pathogen-infected plants exhibit signs of whitish raised spots on the underside of the leaf that enlarge overtime (6 days) and form brownish uredenia (Souza et al., 2008). Uredenia of the pathogen spreads rapidly from 10–22 days post-infection (vegetative—flowering stages), depending on the conditions of the environment (Devi et al., 2020).
Rust races 1, 3, 5 and others were first characterised in other parts of the world (Brazil and United States) and were later observed in Southern Africa, indicating prevalence of numerous races of U. appendiculatus (Liebenberg et al., 2005; Aruga et al., 2012). South Africa’s major production areas including Mpumalanga, Free State, North West, Gauteng and KwaZulu-Natal are greatly affected by rust races 1, 3 and 11. Although rust race 11 is not highly prevalent, it may cause greater yield losses when compared to other races (Liebenberg and Pretorius, 2004). The overall yield of common bean genotypes can be determined by the development of physiological characteristics (Rosales et al., 2012). Any interference by stressors such as diseases at critical phenological stages such as the vegetative development stages of the plant (V1–V4), pre-flowering (V5) and flowering (V6) among others can result in reduced yield (Singh and Schwartz, 2010; Odogwu et al., 2017). Therefore, it is important to utilise an array of methods to manage the U. appendiculatus pathogen in common bean during these critical stages production areas (Souza et al., 2013).
The management of the pathogen relies primarily on the following strategies: i) application of cultural practices, ii) fungicide or chemical application, iii) biological control and iv) host plant resistance (Obongoya et al., 2010). However, due to the aggressiveness of the pathogen and its constant evolution, controlling the pathogen has been problematic (Breiing et al., 2021). Understanding the molecular and biochemical mechanisms involved in plant-host interaction can aid in the development of efficient pathogen management strategies that can improve productivity (Kalavacharia et al., 2000). Progress in legume improvement strategies has been witnessed through the advancement of next-generation sequencing techniques (NGS) and other high throughput genotyping technologies that have been applied to interrogate plant-host interactions (Rubiales et al., 2011; Tayeh et al., 2015; Janila et al., 2016, 2016). This development has led to more omics studies of common bean in response to U. appendiculatus attack (Cooper et al., 2007; Puthoff et al., 2008). For example, methylated and acetylated histones linked to resistance to U. appendiculatus were reported in a genome-wide profiling of histone modifications and gene expression in common bean (Ayyappan et al., 2015). In a similar study, U. appendiculatus resistant genotypes were reported to mediate antioxidant enzymes, phenolic compounds, and other defence genes in response to rust infection (Omara et al., 2022). However, metabolomic changes in common bean after infection with U. appendiculatus have not been addressed yet.
Metabolomic techniques have played a vital role in aiding researchers to identify significant metabolites that contribute to legume improvement (Ramalingam et al., 2015). Profiling of metabolites using LC-MS techniques has been widely utilised to evaluate legume performance under disease pressure (Makhumbila et al., 2022). In a recent study, common bean infected with Fusarium solani significantly enriched biosynthesis of amino acids, flavonoid biosynthesis, purine metabolism and other pathways as a pathogen adaptation strategy (Chen et al., 2020). Amino acids and sugars are among metabolite classes that have been found to be up/down regulated in pea infected with Rhizoctonia solani (Turetschek et al., 2017). Similar results have been observed in common bean infected with Fusarium oxysporum where numerous metabolites were highly enriched after pathogen attack (Chen et al., 2019). Genes of U. appendiculatus have been profiled to efficiently understand the genomic characteristics of the pathogen and it was found that the pathogen alters its genes at different growth stages of the plant (Link, 2020). Although the mode of pathogen action has been evaluated, there is a vast knowledge gap on how common bean genotypes respond to U. appendiculatus infection at different growth stages. In order to understand the dynamics of breeding for rust resistance in common bean, metabolomics can play a vital role in selecting parental genotypes for breeding programmes by providing a basis for resistance biomarkers. In this study, the aim was to evaluate metabolomic changes that occur in resistant (Teebus-RR-1) and susceptible (Golden Gate Wax) common bean genotypes when infected with pathogenic U. appendiculatus fungal races (race 1 and 3) at two-time points (14- and 21- dpi). The functions of the identified metabolites expressed were studied, including their significance to U. appendiculatus tolerance at different growth stages.
2 Materials and methods
2.1 Plant material and treatments
Seeds of common bean genotypes Teebus-RR-1 (resistant/tolerant) and Golden Gate Wax (susceptible) mapped for rust resistant genes (Ur-3 and Ur-6) were obtained from Agricultural Research Council–Grain Crops Institute (ARC-GCI), Potchefstroom, South Africa. Seeds were surface sterilised using 50% bleach solution (Lindsey et al., 2017), rinsed with sterile water, and grown in 9 cm pots with sterile 30 dm3 seedling mix having 50% topsoil and compost, and covered with a vermiculite layer (Figures 1A, B). Inoculation of the respective genotypes was conducted when the leaves were ±
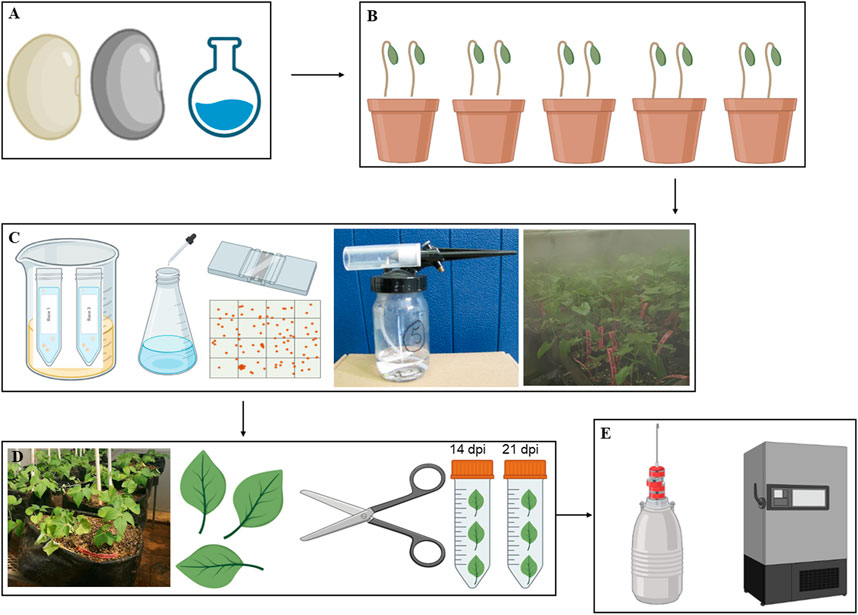
FIGURE 1. A graphical representation of the methodology workflow from seed treatment and planting (A and B), pathogen preparation and inoculation (C), leaf sample harvesting (D) and the storage of harvested material (E).
2.2 Fungal material and inoculation
Spores of U. appendiculatus races 1 and 3 were provided by ARC-GCI (South Africa) for inoculation purposes. The rust races were previously characterised and collected from isolated common bean infected plants, purified, and were stored in a −80°C ultra-freezer. Purified isolates were then re-hydrated by incubating open cryotubes with the pathogen in a glass beaker with warm water and vermiculite (Figure 1B). The beaker containing the cryotubes with the rust pathogens was sealed with sterile cling wrap plastic and left for 12 h at ±18°C. An aqueous soap suspension of Tween 20 (pure liquid soap) with 5 drops per litre of tap water (rust requires rust Ca2+ and Mg2+ ions) was prepared and the rust in cryotubes mixed thoroughly with the suspension. The concentration of rust spores was adjusted to 2.5 × 104 spores per ml using a hemacytometer (Figure 1C) with spore counts repeated four times to obtain 100% germination of spores (Montejo Dominguez et al., 2022).
Plants were spray inoculated on the leaves (the underside leaf targeted) at a low pressure using a compressor attached to a bottle with a spray gun in a disinfected confined booth. The inoculated plants were then left to dry for about 20–30 min and later placed in a dew chamber (Figure 1C) with 95%–100% relative humidity and temperatures of ±18–± 20°C for 48 h. The control experimental plants were mock inoculated with distilled water and subjected to the same treatment as the rust inoculated experimental plants. The inoculated plants were kept in different greenhouse compartments with 28/14°C day/night temperatures with relative humidity of 75% (Singh and Gupta, 2019). The plants were transferred from the 9 cm pots to 50 L black planting bags (Figure 1D) with sterile oxidic soil. The scoring of rust infection severity based on pustule size (Table 1) was conducted at 14- and 21- dpi as described by Hurtado-Gonzales et al. (2017).
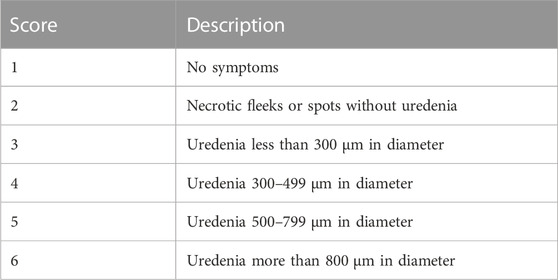
TABLE 1. Uredenia leaf score criteria used for scoring U. appendiculatus severity in common bean plants after inoculation.
2.3 Harvesting and metabolite extraction
Infected (race 1 and 3) and non-infected (controls) leaf material was harvested from the two genotypes at 14- and 21-dpi, representing flowering and pre-flowering stages, respectively (Dann and Deverall, 1996; Devi et al., 2020). The harvested samples were snap frozen with liquid nitrogen and stored in a −80°C ultra-freezer prior to further analysis (Figure 1E). Leaf samples were then weighed (20 mg) and ground into powder in liquid nitrogen using mortar and pestle and extracted using the methanol extraction method consisting of 1.5 mL (1:75 m/v) of 70% LC/MS grade methanol (Merck, Darmstadt, Germany). The extracted samples were vortexed for 30 s, sonicated for 10 min and centrifuged for 5 min at 5,100 rpm (Thermo Fisher, Johannesburg, South Africa). The supernatant was collected and filtered using nylon filters (0.22 µm) into glass vials containing 500 µL inserts (Agela Technologies, Tianjin, China). Three replicates per sample group were prepared for analysis and extracts were stored at 4°C prior to metabolite profiling.
2.4 LC-MS metabolite analysis
Infected (race 1 and 3) and non-infected common bean leaf extracts were subjected to analysis on a liquid chromatography-quadrupole time-of flights tandem mass spectrometry instrument (LCMS-9030 qTOF, Shimadzu Corporation, Kyoto, Japan) for quantification of metabolites at different time intervals. A Shim-pack Velox C18 column (100 mm × 2.1 mm with a 2.7 µm particle size) was used for chromatographic separation at 55°C (Shimadzu Corporation, Kyoto, Japan). An injection volume of 3 µL was used for all samples and were run on a binary mobile phase including solvent A: 0.1% formic acid in Milli-Q HPLC grade water (Merck, Darmstadt, Germany) and solvent B: UHPLC grade methanol with 0.1% formic acid (Romil Ltd., Cambridge, United Kingdom). Chromatographic analysis was done using qTOF high-definition mass spectrometer that was set to negative electrospray ionisation for data acquisition. Parameters set included nebulization, interface voltage (4.0 kV), interface temperature (300°C), dry gas flow (3 L/min), detector voltage (1.8 kV), heat block (400°C), DL (280°C) and flight tube (42°C) temperatures. Ion fragmentation was achieved using argon gas for collision with an energy of 30 eV and 5 eV spread (Ramabulana et al., 2021).
2.5 Multivariate data analysis
Data pre-processing was done using XCMS, with HPLC/UHD-qTOF parameters using the centWave feature detection method, maximum threshold of 15 ppm, a signal to noise ratio of 6, prefilters for intensity and noise at 100 and 3. The retention time correction method was obiwarp with profStep, while the alignment minimum fraction of samples was 0.5 and a 0.015 m/z width. Kruskal–Wallis statistical test was applied to the data that resulted in a feature table with 11,315 characteristics. The feature table was exported to SIMCA version 17.0 software, normalised and pareto scaled prior to model application. Principal Component Analysis (PCA) and Orthogonal Projection to Latent Structures—Discriminant Analysis (OPLS-DA) models were applied to the data.
2.6 Metabolite annotation, relative quantification and pathway analysis
MzMine v2.3 was used for data visualisation, chromatogram deconvolution, MS1/MS2 building, isotope grouping, alignment, filtering and gap filling (Pluskal et al., 2010). The resulting mascot generic format (mfg.) file and metadata for the respective treatments were processed on GNPS online. Libraries used for spectral search included GNPS, ChEBI, HMBD, DRUGNANK, FooDB, and SUPNAT (Wang, 2016). Metabolites were matched to GNPS linked databases and were also putatively annotated or verified through searches in compound databases using their peak mass and isomeric SMILES. Databases including KEGG compound, KNApSAcK, Chemspider, ChEBI, PubChem, and Dictionary of Natural Products. Annotations were further confirmed through literature search of related studies. Metabolite concentrations were used for an overview of metabolomic pathways that were enriched. Overrepresentation with a hypergeometric test and KEGG metabolite pathway for Arabidopsis thaliana (thale cress) was used for pathway analysis in MetaboAnalyst v5.0.
3 Results
3.1 Phenotypic evaluation of common bean post inoculation with U. appendiculatus
Plant leaves of Teebus-RR-1 and Golden Gate Wax common bean genotypes were evaluated phenotypically for their response to U. appendiculatus race 1 and 3 throughout the experiment (Figure 2; Table 1). The resistant genotype Teebus-RR-1 exhibited no symptoms of infection by race 1 and 3 at 14 dpi (Figures 2B, C). Similar phenotypic observations were observed at 21 dpi on secondary leaves, after the primary leaves had matured and fallen off (Figures 2E, F). Golden Gate Wax had the highest number of pustules when infected with rust races 1 and 3 at the two time points post infection (Figures 2H, I). Secondary leaves of the susceptible genotypes exhibited a continuous spread of infection with rust race 1 at 21 dpi (Figure 2K) while race 3 had no visible pustules (Figure 2L). Leaf lesions, necrosis and wilting were among symptoms that were prevalent on susceptible genotype Golden Gate Wax at 14 and 21 dpi compared to the control mock inoculated leaves (Figures 2G, J).
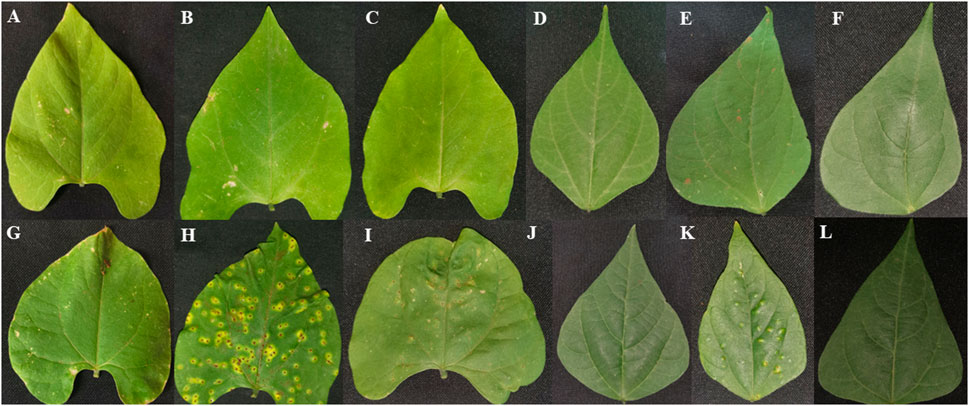
FIGURE 2. Phenotypic leaf evaluation of Teebus-RR-1 and Golden Gate Wax genotypes in response to U. appendiculatus infection. (A) Teebus-RR-1 control mock inoculated at 14 dpi, (B) Teebus-RR-1 race 1 inoculated leaf at 14 dpi, (C) Teebus-RR-1 race 3 inoculated leaf at 14 dpi, (D) Teebus-RR-1 control mock inoculated at 21 dpi, (E) Teebus-RR-1race 1 inoculated at 21 dpi and (F) Teebus-RR-1 race 3 inoculated at 21 dpi. (G) Golden Gate Wax control mock inoculated at 14 dpi, (H) Golden Gate Wax race 1 inoculated at 14 dpi, (I) Golden Gate Wax race 3 inoculated leaf at 14 dpi, (J) Golden Gate Wax control mock inoculated at 21 dpi, (K) Golden Gate Wax race 1 inoculated at 21 dpi and (L) Golden Gate Wax race 3 inoculated at 21 dpi.
3.2 Comprehensive analysis of common bean metabolites
Untargeted metabolite profiles of genotypes Teebus-RR-1 and Golden Gate Wax were analysed using LC-MS at 14- and 21- dpi in response to U. appendiculatus. The Principal Component Analysis (PCA) model provided the virtual analysis of the effects of U. appendiculatus treatments on common bean, revealing clustering on genotypes, races and time intervals post infection. The PCA results showed clustering between the two genotypes (Supplementary Figure S1). Although the PCA score plot showed differential sample clustering with a separation of genotypes (indicating differential metabolite profiles), Othorgonal (OPLS-DA) was computed to allow prediction of variations, consequently allowing identification of potential biomarkers (Tugizimana et al., 2013). The OPLS-DA results revealed similar clustering patterns between the two genotypes (Supplementary Figure S2), reflecting the differences in metabolite profiles between the genotypes.
A total of 71 known metabolites were identified to be present in both genotypes at varying concentrations for the different treatments and a heatmap with the 33 metabolites were found to be significant (Table 2). Interestingly, excessive metabolite changes were observed between treatments of the susceptible genotype Golden Gate Wax compared to the resistant genotype Teebus-RR-1 that had slight or limited metabolite changes when subjected to U. appendiculatus infection at the different time points. An example, afzelechin-(4alpha->8)-afzelechin, (5-Phenyl-1,2,4-triazol-3-yl) urea, tuberonic acid glucoside, xanthotoxin, chlorflavonin, D-sucrose and linoleate were highly concentrated in Teebus-RR-1 samples infected with rust race 1 at 14 dpi (T114) while there were low concentrations of these metabolites in Golden Gate Wax samples (G114) under similar conditions. These metabolites were further expressed in lower concentrations at 21 dpi race 1 samples (G121) while the resistant genotype kept a moderately high production of these metabolites in samples (T121) at 21 dpi. Kaempferol 3-O-rhamnoside-7-O-glucoside, phylloquinone and sennoside D were produced in moderately high concentrations in the resistant genotype Teebus-RR-1 at both 14 and 21 dpi in race 3 infected samples (T3) while these metabolites were suppressed in the susceptible genotype Golden Gate Wax race 3 infected samples (G3) at both time points (Figure 3). The majority of the differentially expressed metabolites in both these cultivars belong to an array of compound classes including flavonoids, terpenoids, fatty acids and phenols that have been found in numerous plant species including legumes (Table 2; Supplementary Table S1).
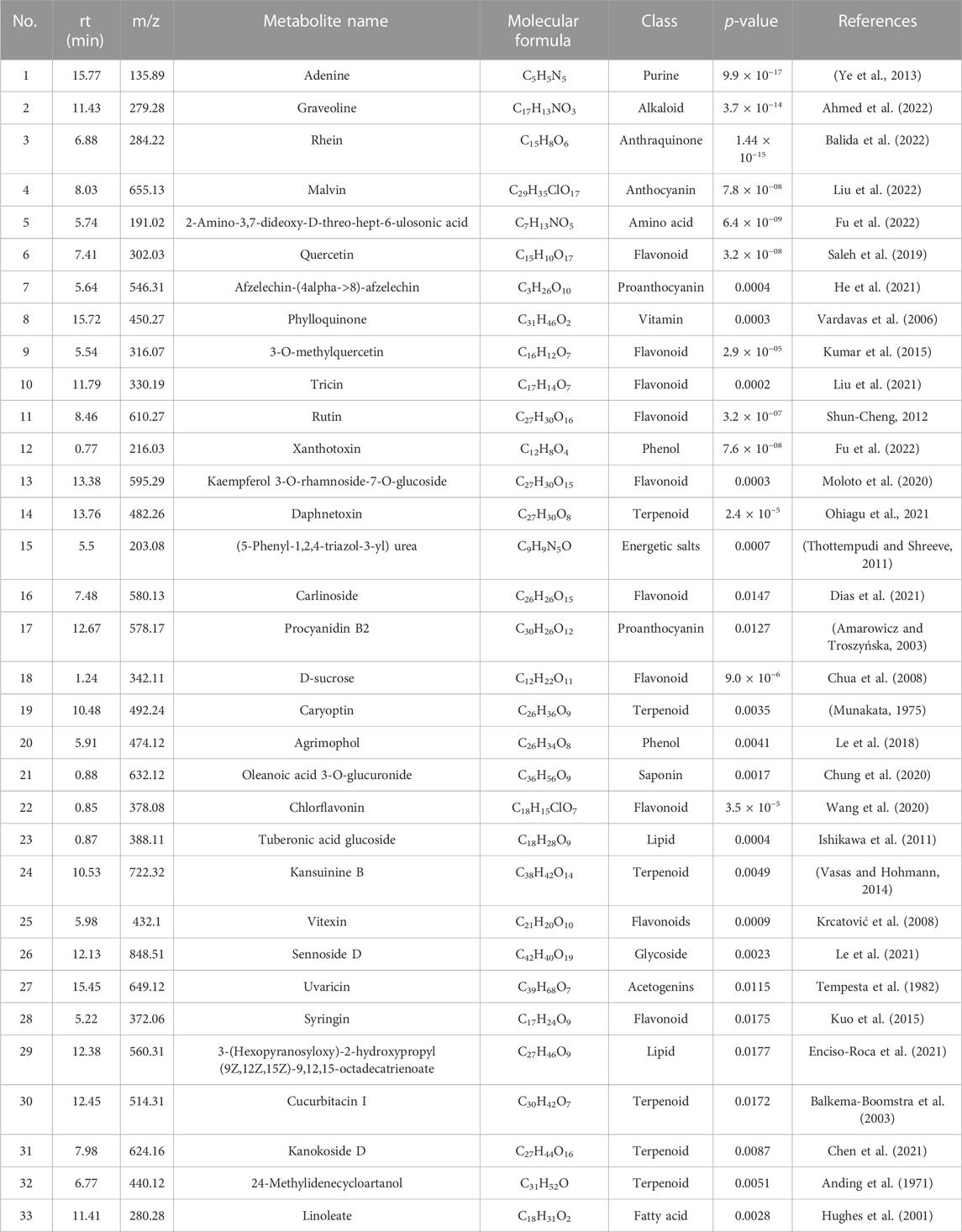
TABLE 2. Putatively identified metabolites that were significantly expressed in common bean genotypes infected with U. appendiculatus strains.
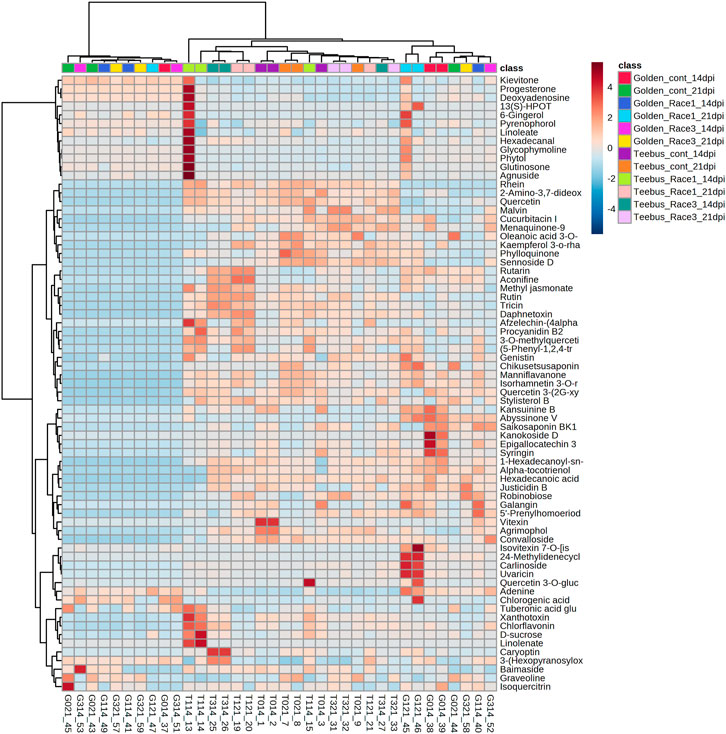
FIGURE 3. Heatmap of metabolite concentrations of common bean leaves of resistant genotype Teebus_RR_1 and susceptible genotype Golden Gate Wax infected with U. appendiculatus races 1 and 3 at two time points (14- and 21- dpi). Class colours represent the experimental treatments corresponding to samples, while numbers show the up/down regulation metabolites identified.
The resistant genotype Teebus-RR-1 enriched more metabolites in rust infected plants compared to non-infected control samples (Figure 3) for the different treatments. An upregulation of metabolites was prevalent in the resistant genotype Teebus-RR-1 compared to the susceptible genotype that had minimum regulation of metabolites leading to fewer being significant (Figures 5A–C). Metabolites belonging to compound classes such as flavonoids including D-sucrose, 3-O-methylquercetin, chlorflavonin and phenol xanthotoxin (Figure 4A) were enriched in U. appendiculatus race 1 infected samples at 14 dpi; while vitexin, isovitexi-7-O glucoside, saikosaponinBK1 and agrimophol were down regulated (Figures 4A–C). Other metabolites belonging to an array of compound classes such as lipids, anthocyanins and energetic salts were also differentially enriched in the resistant genotype Teebus-RR-1 at 14 dpi when infected with U. appendiculatus race 1 (Figures 4D,F; Table 2). At 21 dpi, most of the differentially expressed metabolites including flavonoids and other compound classes were up regulated and expressed at exponentially higher concentrations, except for phylloquinone (Figures 4E, F). On the other hand, quercetin, galangin and rutin were up regulated, while vitexin was down regulated in the Teebus-RR-1 infected with race 3 U. appendiculatus pathogen at 14 dpi (Figure 4G). Furthermore, terpenoids daphnetoxin, and kansuinine B were differentially expressed at varying concentrations when compared to the control (Figure 4H). In the resistant genotype Teebus-RR-1, flavonoids and terpenoids were expressed at slightly higher concentrations in the U. appendiculatus race 3 infected samples at 21 dpi in comparison to control samples (Figures 4I, J).

FIGURE 4. Box plots illustrating concentrations of metabolites differentially enriched in resistant genotype Teebus-RR-1 in response to U. appendiculatus race 1 at 14 dpi (A–D), 21 dpi (E–F), race 3 at 14 dpi (G and H) and 21 dpi (I and J). (A) Flavonoids. (B) Terpenoids. (C) Phenols. (D) Other metabolites belonging to an array of compound classes. (E) Flavonoids. (F) Other metabolites belonging to an array of compound classes. (G) Flavonoids. (H) Terpenoids. (I) Flavonoids. (J) Terpenoids. The red indicates the peak area quantification of metabolites extracted from the control and green indicates the peak area quantification of metabolites extracted from plants infected with U. appendiculatus.
In the susceptible genotype Golden Gate Wax, isovetexin-7-glucoside was down regulated at 14 dpi with U. appendiculatus race 1 (Figures 5A,D), while graveoline and tuberonic acid glucoside were also down regulated at 21 dpi (Figures 5B, C, E–G). There were no differentially expressed metabolites at 14 dpi with U. appendiculatus race 3 (Supplementary Figure S3). Meanwhile, tuberonic acid glucoside was differentially down regulated at 21 dpi in U. appendiculatus race 3 infected plants (Figure 5C).
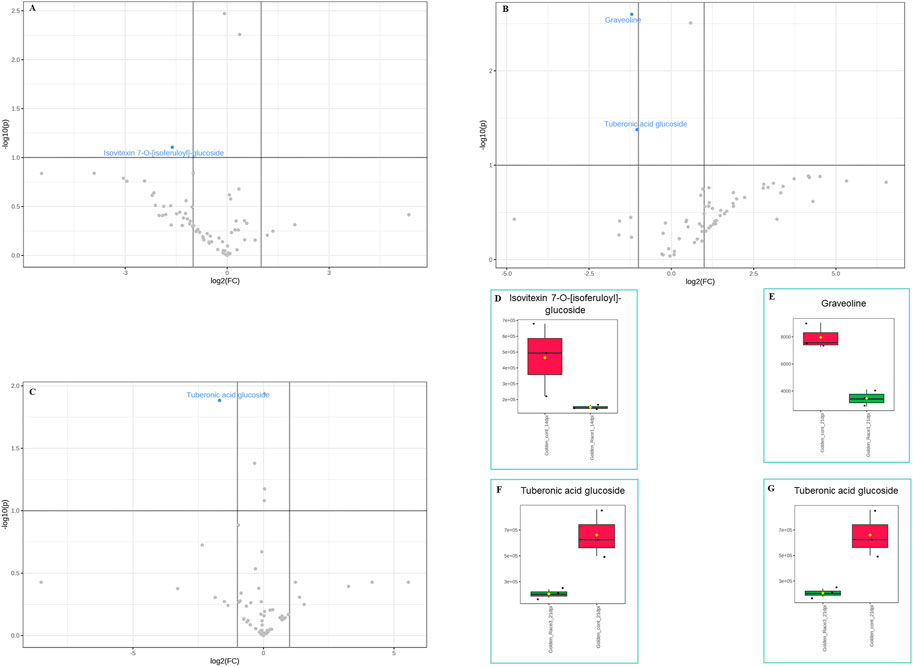
FIGURE 5. Volcano and box plots showing metabolite changes and concentrations in common bean susceptible genotype Golden Gate Wax response to U. appendiculatus race 1 at 14 dpi (A&D—flavonoid), 21 dpi [B, (E) alkaloid and (F) lipid], race 3 at 21 dpi (C&G—lipid). The blue indicates significantly down regulated metabolites while grey indicates non-significant metabolites in U. appendiculatus infected plants. The red indicates the peak area quantification of metabolites extracted from the control and green indicates the peak area quantification of metabolites extracted from plants infected with U. appendiculatus.
3.3 Infection with U. appendiculatus triggers defence metabolomic pathways
The KEGG pathway analysis revealed that numerous metabolites that were putatively annotated were associated with several pathways including alpha-linolenic acid metabolism, biosynthesis of fatty acids, flavonoid biosynthesis and purine metabolism (Figure 6). Alpha-linolenic acid metabolism and biosynthesis of unsaturated fatty acids pathway are known to synthesise a group of fatty acids including linolenate, methyl jasmonate, hexadecenoic acid and linoleate. Linolenate is known as a backbone of metabolite 12(S)-HPOT. Meanwhile, flavonoid biosynthesis pathway resulted in the biosynthesis of quercertin from which 3-O-methylquercetin and quercetin 3-O-glucuronide were derived (Figure 6; Supplementary Table S1). Purine metabolism pathway on the other hand was also present and yielded adenine and deoxyadenosine.
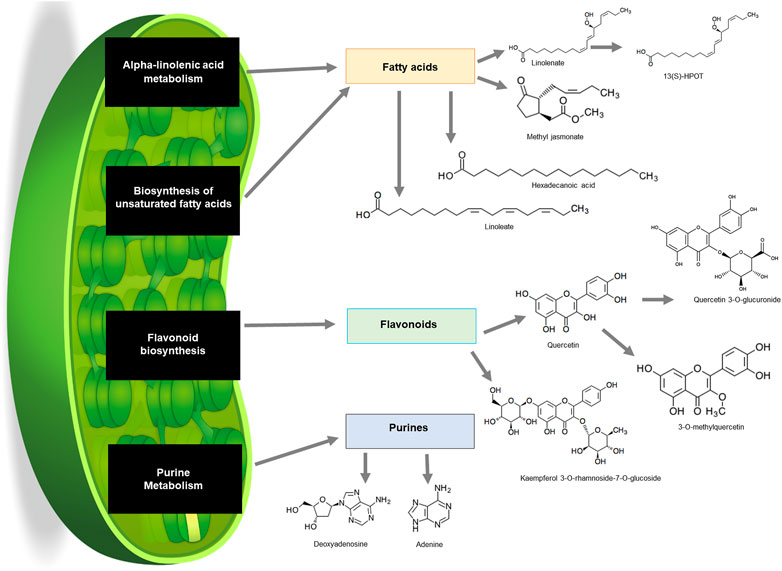
FIGURE 6. Schematic summary of pathways in common bean plants that contributed to the production of a wide variety of compounds from different classes on KEGG. Fatty acids (orange), flavonoids (green) and purines (blue) from leaf samples.
4 Discussion
4.1 Metabolomic analysis of P. vulgaris genotypes in response to U. appendiculatus
Metabolites are products that play major roles in plant response to environmental stress conditions. Currently, there is limited knowledge on metabolites involved in resistance against an array of fungal and bacterial pathogens in legumes (Makhumbila et al., 2022). In this study, an untargeted metabolome profiling of two common bean genotypes Teebus-RR-1 and Golden Gate Wax were investigated for their response to race 1 and 3 U. appendiculatus infections at different time-points (14 and 21 days). The analysis demonstrated that under U. appendiculatus infection, different compounds were enriched at varying concentrations in both genotypes at different time points. The resistant genotype showed dominance in metabolite expression in response to U. appendiculatus infections when compared to non-infected controls. For instance, 3-O-methylquercetin was highly enriched at 14 dpi when treated with race 1 in the resistant genotype. An increase in 3-O-methylquercetin has been associated with an increase of hydrogen peroxide (H2O2)) due to biotic stress conditions, which is converted from reactive oxygen species (Quan et al., 2008; Kumar et al., 2015). H2O2 is a signalling molecule that regulates development and stress adaptation in plants. In addition, Singh et al. (2021) reported that an increase of 3-O-methylquercetin can also be attributed to an increased accumulation of quercetin in resistant genotypes under biotic stress. Quercetin is a flavonoid known to play a role in the process of protecting plants against stress effects. Moreover, quercetin and rutin were also found to be upregulated in the resistant genotype at 21 dpi. Rutin is a flavonoid that plays a key role in protecting plants against pathogens (Kianersi et al., 2020). The increase in flavonoid levels such as quercetin and its derivatives, and rutin among others, have been reported in pathogen-infected plants (Hadrami et al., 2011; Kianersi et al., 2020). In this study, similar results were observed in the Teebus-RR-1 genotype, suggesting the role of flavonoids in defence in response to U. appendiculatus at different time-points.
Terpenoids such as daphnetoxin was induced in the resistant genotype at 14 dpi compared to the non-infected control. Daphnetoxin has been reported for its toxicity (Chen et al., 2022), possibly contributing to the suppression of the pathogen growth in the plant. On the other hand, kansuinine B was suppressed in the same genotype at 14 dpi compared to non-infected control. Reduced concentrations of terpenoids have been reported in Chrysanthemum morifolium inoculated with Alternaria tenuissima (He et al., 2022). The decreased concentration of terpenoids have been shown to cause cell membrane damage as they merge with phosphor-lipid acyl chains (Hammerbacher et al., 2019). In addition, an alkaloid such as aconifine was also moderately enriched in a resistant U. appendiculatus treated genotype (Figure 3). Generally, alkaloids have been reported for their role in herbivory defence in plants (Macel, 2011; Bhambhani et al., 2021), suggesting their possible role in common bean response to U. appendiculatus.
On the other hand, the susceptible Golden Gate Wax genotype differentially expressed compounds such as alkaloids and lipids at different time-points post infection with rust races 1 and 3. Metabolites that were downregulated in the susceptible genotype in response to U. appendiculatus races included tuberonic acid glucoside (Figure 5). The downregulation of lipids can be linked to lipid metabolism degradation in plants infected with pathogens, therefore signalling the genotypes’ susceptibility (Robison et al., 2018; Foucher et al., 2020). In addition, U. appendiculatus slows down photosynthetic activity by forming pustules on susceptible genotypes, as it was observed in our study (Figure 2H), consequently causing the cell wall to collapse by lipid peroxidation (Bojtor et al., 2019). An exploration of other metabolites in the susceptible genotype indicated a dramatic decrease in the expression of phenolic compounds (xanthotoxin and chlorogenic acid), which are essential in the plants defence against U. appendiculatus (Omara et al., 2022). The pathogen U. appendiculatus could be promoting the synthesis of proteins for feeding that are synthesised from an array of pathways including lipids (Link, 2020). This group of metabolites were downregulated in the susceptible genotype in our study.
It has been widely reported that exposure of plants to pathogen stress increases the production of flavonoids, terpenoids and phenols as a defence strategy (López-Gresa et al., 2010). Flavonoids signal compounds in plant-pathogen interaction that also tend to be highly produced in resistant as compared to susceptible genotypes (Steinkellner et al., 2007; Chin et al., 2018). Terpenoids influence the plant’s ability to inhibit pathogen attack (Toffolatti et al., 2021). Phenols protect plant tissues from the toxic effects of pathogens (Kumar et al., 2015), while alkaloids store nitrogen reserves during pathogen attack (Ali et al., 2019).
The results of this study also show that due to U. appendiculatus infection, compounds such as D-sucrose, 3-O-methylquercetin, phenol xanthotoxin and chlorflavonin were highly enriched in the resistant genotype compared to the susceptible genotype. A recent study reported similar findings that resistant genotypes tend to secrete a larger number of metabolites at high concentrations when compared to susceptible genotypes under disease pressure (Robison et al., 2018). Additionally, in barley genotypes infected with stripe rust, similar patterns of metabolite upregulation in resistant genotypes were reported (Singla et al., 2020).
4.2 Defence strategies of common bean plants to U. appendiculatus
In this study, we found that chlorflavonin, D-sucrose, xanthotoxin, tuberonic acid glucoside, malvin, vitexin, robinobiose, 3-O-methylquercetin, rutarin, aconifine, galangin, rutin, daphnetoxxin, syringin and 24-Methylidenecycloartanol were highly enriched metabolites in response to U. appendiculatus infection with aconifine being the most enriched (Figure 4F). Aconifine is an alkaloid that has been widely linked to Aconitum karakolicum Rapaics which is an antiproliferative that prohibits pathogen cell growth (Tashkhodjaev and Sultankhodjaev, 2009; Thawabteh et al., 2019). On the other hand, rutarin was also expressed in abundance at 21 dpi when the plant was under rust infection. The compound has been reported to have antifeedant/antiparasitic properties in tree plants (Lacroix et al., 2011). Flavonoid D-sucrose was also enriched in common bean at 14 dpi in the resistant genotype. Certain studies have highlighted the importance of this metabolite in plant growth and associated the metabolite with fungal pathogen virulence in maize (Wahl et al., 2010). In another study, it was highlighted that the synthesis of sucrose is beneficial for the plants overall growth and tolerance to stressors (Stein and Granot, 2019). Xanthotoxin, a phenolic compound was also differentially expressed in plants infected with the U. appendiculatus pathogen. This compound has been appraised for its ability to interact with free radicals and biological molecules that might cause lipid peroxidation, protein damage, enzyme inhibition and genetic oxidation that might result in cell death (Bajerová et al., 2014; Dumanović et al., 2021). Another antioxidant characterized compound that was differentially expressed was galangin that has been found to be a beneficial antioxidant in red kidney bean evaluated under water deficiency (Manoj et al., 2022). This is in concurrence with our finding that certain metabolites are highly enriched in common bean leaves as a stress defence strategy. However, it is still unclear how metabolites interact with genes expressed, and for this reason, an integrated omics study to unravel underlying response mechanisms involved in common bean rust interactions will have far reaching impact in common bean breeding.
4.3 Metabolomic pathways contribute to the plants ability to respond to U. appendiculatus attack
Fatty acids that are synthesized by the plant tend to undergo prokaryotic or eukaryotic pathway. During this occurrence, they are converted to complex lipids for assembly in the eukaryotic pathways. This consequently leads to the production of plant fats that contribute to the plants overall nutrition (Chellamuthu et al., 2022). In the current study, metabolites synthesized through the alpha-linolenic acid metabolism and biosynthesis of unsaturated fatty acids were not significantly enriched. This possibly indicates that despite stress conditions, other metabolomic activities responsible for the overall productivity still occur. Interestingly, Kumaraswamy et al. (2011) reported a 4-fold expression of linolenic acid in barley resistant genotypes infected with Fusarium graminearum compared to the susceptible genotypes.
In terms of flavonoids, they have been widely reported for their contribution in plant biotic and abiotic stress response (Jan et al., 2022). Our results found that quercetin, rutin and galangin were up regulated in the resistant genotype infected with U. appendiculatus at both time points. Similar findings reported quercetin and kaempferol derivatives to be effective contributors to plants stress response because of their antioxidant properties (Koskimäki et al., 2009). The expression of flavonoids in the susceptible genotype (Golden Gate Wax) was lower when compared to the resistant, suggesting reduced flavonoid biosynthesis. This is supported by the study of Nagamatsu et al. (2007) who reported that reduced flavonoid biosynthesis in soybean under viral pathogen stress may be associated with cell wall degradation. Similarly, purine metabolism has also been appraised for its contribution to stress adaptation by contributing to nitrogen metabolism (Watanabe et al., 2014). However, metabolites belonging to the purine metabolism pathway were not significantly enriched in the current study. The biosynthesis of metabolites in plants is related to genes (Raza, 2020) and thus complex, therefore metabolomics and transcriptomics studies should be integrated to gain insight on the integral pathways that play a role in defence.
5 Conclusion
In the present study, the whole metabolome of common bean infected with U. appendiculatus was characterised. The differences in terms of metabolites between the inoculated and non-inoculated common bean showed that flavonoids, terpenoids, alkaloids and others play an important role in defence response to U. appendiculatus infection. Flavonoid pathway was the main defence response in common bean. Further investigations on early response (24–72 h post-infection) will give a clearer picture of the metabolites involved at the initial stages of infection. Metabolomic expression patterns of genotypes at early and late infection stages can be used to understand plant-pathogen interactions and significantly upregulated metabolites can serve as biomarkers in breeding programmes. Overall, this study provides new insights in understanding common bean interactions with U. appendiculatus. Future research studies should focus on integrating metabolomics with other OMICs to uncover possible underlying defence mechanisms which could represent a helpful tool for developing common bean resistant varieties toward U. appendiculatus.
Data availability statement
Metabolomic data presented in the study are deposited in the MetaboLights repository, accession number MTBLS6972.
Author contributions
SF, MR, and HM conceptualised and designed the experiment. PM conducted the plant growth experiments and applied the treatments. MR, SF, and PM conducted the metabolite extractions. PM, MR, and NM conducted the LC-MS analysis. Data analysis was done by PM, MR, and SF. Revision of the manuscript was by PM, MR, HM, NM, and SF.
Funding
This work was supported by the University of South Africa’s Women in Research Support Programme together with the National Research Foundation (MND2104145947060). The funders had no role in the study design, data collection and analysis. The decision to publish the prepared manuscript was solely by the authors.
Acknowledgments
We would like to acknowledge ARC-GCI for their provision of experimental facilities and germplasm.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1141201/full#supplementary-material
SUPPLEMENTARY TABLE S1 | A list of all putatively identified differentially expressed metabolites expressed across common bean groups evaluated under U. appendiculatus stress.
SUPPLEMENTARY FIGURE S1 | The PCA score plot of P. vulgaris genotypes with varying resistance to U. appendiculatus races.
SUPPLEMENTARY FIGURE S2 | The OPLS- DA scores plot of P. vulgaris genotypes with varying resistance to U. appendiculatus races.
SUPPLEMENTARY FIGURE S3 | Volcano plot showing none significant metabolites in susceptible genotype Golden Gate Wax infected with U. appendiculatus race 3 at 14 dpi.
References
Ahmed, S. M., Kumar, A., and Gandhi, S. G. (2022). Exogenously applied rohitukine inhibits photosynthetic processes, growth and induces antioxidant defense system in Arabidopsis thaliana. Antioxidants 11, 1512. doi:10.3390/antiox11081512
Ali, A., Khan, A., Sharif, R., Mujtaba, M., and Gao, S. J. (2019). Sugarcane omics: An update on the status of research and crop improvement. Plants 8 334. doi:10.3390/plants8090344
Alleyne, A. T., Steadman, J. R., and Eskridge, K. M. (2008). Monitoring changing virulence patterns of Uromyces appendiculatus in the resistant pinto bean cultivar olathe by rep-PCR. Eur. J. Plant Path 122, 315–319. doi:10.1007/s10658-008-9295-0
Amarowicz, R., and Troszyńska, A. (2003). Antioxidant activity of extract of pea and its fractions of low molecular phenolics and tannins. Pol. J. Food Nutr. Sci. 53, 10–15.
Anding, C., Brandt, R. D., and Ourisson, G. (1971). Sterol biosynthesis in Euglena gracilis z. sterol precursors in light-grown and dark-grown. Euglena gracilis z. Eur. J. Biochem. 24, 259–263. doi:10.1111/j.1432-1033.1971.tb19679.x
Bajerová, P., Adam, M., Bajer, T., and Ventura, K. (2014). Comparison of various techniques for the extraction and determination of antioxidants in plants. J. Sep. Sci. 37, 835–844. doi:10.1002/jssc.201301139
Balida, L. A. P., Theresa, J., Regalado, A., Teodosio, J. J. R., Dizon, K. A. H., Sun, Z., et al. (2022). Antibiotic isoflavonoids, anthraquinones, and pterocarpanoids from pigeon pea (Cajanus cajan L.) seeds against multidrug-resistant Staphylococcus aureus. Plant path 12, 279. doi:10.3390/metabo12040279
Balkema-boomstra, A. G., Zijlstra, S., Verstappen, F. W. A., Inggamer, H., Mercke, P. E., Jongsma, M. A., et al. (2003). Role of cucurbitacin c in resistance to spider mite (Tetranychus urticae) in cucumber (Cucumis sativus L.). J. Chem. Ecol. 29, 225–235. doi:10.1023/a:1021945101308
Bhambhani, S., Kondhare, S. R., and Giri, A. P. (2021). Diversity in chemical structures and biological properties of plant alkaloids. Molecules 26, 3374. doi:10.3390/molecules26113374
Bojtor, C. S., Győri, Z., Sipos, P., Radócz, L., and Tóth, B. (2019). Effect of bean rust [ Uromyces appendiculatus (pers.) strauss] on photosynthetic characteristics, superoxide-dismutase activity, and lipid peroxidation of common bean (Phaseolus vulgaris L.). Acta Aliment. 48, 253–259. doi:10.1556/066.2019.48.2.13
Breiing, V., Hillmer, J., Schmidt, C., Petry, M., Behrends, B., Steiner, U., et al. (2021). Fungicidal efficacy of drying plant oils in green beans against bean rust (Uromyces appendiculatus). Plants 10, 143. doi:10.3390/plants10010143
Chellamuthu, M., Kumaresan, K., and Subramanian, S. (2022). Increase in alpha-linolenic acid content by simultaneous expression of fatty acid metabolism genes in sesame (Sesamum indicum). physiology Mol. Biol. plants 28, 559–572. doi:10.1007/s12298-022-01152-0
Chen, H., Singh, H., Bhardwaj, N., Bhardwaj, S. K., Khatri, M., Kim, K., et al. (2022). An exploration on the toxicity mechanisms of phytotoxins and their potential utilities. Crit. Rev. Environ. Sci. Technol. 52, 395–435. doi:10.1080/10643389.2020.1823172
Chen, L., Wu, Q., He, T., Lan, J., Ding, L., Liu, T., et al. (2020). Transcriptomic and metabolomic changes triggered by Fusarium solani in common bean (Phaseolus vulgaris L.). Genes 11, 177. doi:10.3390/genes11020177
Chen, L., Wu, Q., He, W., He, T., Wu, Q., and Miao, Y. (2019). Combined de novo transcriptome and metabolome analysis of common bean response to Fusarium oxysporum f. sp. phaseoli infection. Int. J. Mol. Scie. 20, 6278. doi:10.3390/ijms20246278
Chen, N. W. G., Ruh, M., Darrasse, A., Foucher, J., Briand, M., Costa, J., et al. (2021). Common bacterial blight of bean: A model of seed transmission and pathological convergence. Mol. Plant Path. 22, 1464–1480. doi:10.1111/mpp.13067
Chin, S., Behm, C. A., and Mathesius, U. (2018). Functions of flavonoids in plant–nematode interactions. Plants 7, 85. doi:10.3390/plants7040085
Chua, T. K., Bujnicki, J. M., Tan, T., Huynh, F., Patel, B. K., and Sivaraman, J. (2008). The structure of sucrose phosphate synthase from halothermothrix orenii reveals its mechanism of action and binding mode. Plant Cell 20, 1059–1072. doi:10.1105/tpc.107.051193
Chung, S. Y., Seki, H., Fujisawa, Y., Shimoda, Y., Hiraga, S., Nomura, Y., et al. (2020). A cellulose synthase-derived enzyme catalyses 3-o-glucuronosylation in saponin biosynthesis. Nat. Comm. 11, 5664. doi:10.1038/s41467-020-19399-0
Cooper, B., Neelam, A., Campbell, K., Lee, J., Liu, G., Garrett, W., et al. (2007). Protein accumulation in the germinating Uromyces appendiculatus uredospore. Mol. Plant-Microbe Inter. 20, 857–866. doi:10.1094/MPMI-20-7-0857
Dann, E. K., and Deverall, B. J. (1996). 2, 6-dichloro-isonicotinic acid (ina) induces resistance in green beans to the rust pathogen, Uromyces appendiculatus, under field conditions. Australas. Plant Path 25, 199–204. doi:10.1071/ap96034
Delgado, D. Z., de Freitas, M. B., and Stadnik, M. J. (2013). Effectiveness of saccharin and ulvan as resistance inducers against rust and angular leaf spot in bean plants (Phaseolus vulgaris). Crop Prot. 47, 67–73. doi:10.1016/j.cropro.2013.01.003
Devi, B., Gupta, S. K., Singh, G., and Prasad, P. (2020). Efficacy of new generation fungicides against French bean rust caused by Uromyces appendiculatus. Phytoparasitica 48, 535–543. doi:10.1007/s12600-020-00820-9
Dias, M. C., Pinto, D. C. G. A., and Silva, A. M. S. (2021). Plant flavonoids: Chemical characteristics and biological activity. Molecules 26, 5377. doi:10.3390/molecules26175377
Dumanović, J., Nepovimova, E., Kuča, M. N. K., Jaćević, V., and Jacevic, V. (2021). The significance of reactive oxygen species and antioxidant defense system in plants: A concise overview. Front. Plant Scie. 11. doi:10.3389/fpls.2020.552969
Enciso-roca, E. C., Aguilar-felices, E. J., Tinco-jayo, J. A., Arroyo-acevedo, J. L., and Calderon, O. H. (2021). Biomolecules with antioxidant capacity from the seeds and sprouts of 20 varieties of Chenopodium quinoa willd. (quinoa). Plants 10, 2417. doi:10.3390/plants10112417
Foucher, J., Ruh, M., Préveaux, A., Carrère, S., Pelletier, S., Briand, M., et al. (2020). Common bean resistance to xanthomonas is associated with upregulation of the salicylic acid pathway and downregulation of photosynthesis. BMC Genomics 21, 566–6. doi:10.1186/s12864-020-06972-6
Fu, K., Zhang, J., Wang, l., Zhao, X., and Luo, Y. (2022). Xanthotoxin induced photoactivated toxicity, oxidative stress and cellular apoptosis in Caenorhabditis elegans under ultraviolet A. Comp. Biochem. Physiol. C. Toxicol. Pharmacol. 251, 109217. doi:10.1016/j.cbpc.2021.109217
Gross, P. L., and Venette, J. R. (2001). Overwinter survival of bean rust urediniospores in North Dakota. Plant Dis. 85, 226–227. doi:10.1094/pdis.2001.85.2.226
Hadrami, E., Adam, A. L., and Daayf, F. (2011). Biocontrol treatments confer protection against Verticillium dahliae infection of potato by inducing antimicrobial metabolites. Mol. Plant-Microbe Interact. 24, 328–335. doi:10.1094/MPMI-04-10-0098
Hammerbacher, A., Coutinho, T. A., and Gershenzon, J. (2019). Roles of plant volatiles in defence against microbial pathogens and microbial exploitation of volatiles. Plant, Cell Environ. 42, 2827–2843. doi:10.1111/pce.13602
He, X., Jiang, Y., Chen, S., Chen, F., and Chen, F. (2022). Terpenoids and their possible role in defense against a fungal pathogen Alternaria tenuissima in Chrysanthemum morifolium cultivars. J. Plant Growth Reg., 1–14. doi:10.1007/s00344-022-10619-z
He, X., Yang, F., and Huang, X. A. (2021). Proceedings of chemistry, pharmacology, pharmacokinetics and synthesis of biflavonoids. Molecules 26, 6088. doi:10.3390/molecules26196088
Hughes, R. K., Lawson, D. M., Hornostaj, A. R., Fairhurst, S. A., and Casey, R. (2001). Mutagenesis and modelling of linoleate-binding to pea seed lipoxygenase. Euro. J. Biochem. 268, 1030–1040. doi:10.1046/j.1432-1327.2001.01964.x
Hurtado-Gonzales, O. P., Valentini, G., Gilio, T. A., Martins, A. M., Song, Q., and Pastor-Corrales, M. A. (2017). Fine mapping of Ur-3, a historically important rust resistance locus in common bean. G3 Genes Genomes Genet. 7, 557–569. doi:10.1534/g3.116.036061
Ishikawa, F. H., Ramalho, M. A. P., and Souza, E. A. (2011). Common bean lines as potential differential cultivars for race 65 of. Colletorichum Lindemuthianum. J. Plant Path. 5, 461–464.
Jan, R., Khan, M., Asaf, S., LubnaAsifKim, S. K. M., and Kim, K. M. (2022). Bioactivity and therapeutic potential of kaempferol and quercetin: New insights for plant and human health. Plants 11, 2623. doi:10.3390/plants11192623
Janila, P., Variath, M. T., Pandey, M. K., Desmae, H., Motagi, B. N., Okori, P., et al. (2016). Genomic tools in groundnut breeding program: Status and perspectives. Front. Plant Scie. 7, 289. doi:10.3389/fpls.2016.00289
Kalavacharia, V., Stavely, J. R., Myers, J. R., and McClean, P. E. (2000). crg, a gene required for ur-3 -mediated rust resistance in common bean, maps to a resistance gene analog cluster. Mol. Plant Microbe Interact. 13, 1237–1242. doi:10.1094/MPMI.2000.13.11.1237
Kianersi, F., Abdollahi, M. R., Mirzaie-Asl, A., Dastan, D., and Rasheed, F. (2020). Identification and tissue-specific expression of rutin biosynthetic pathway genes in Capparis spinosa elicited with salicylic acid and methyl jasmonate. Sci. Rep. 10, 8884. doi:10.1038/s41598-020-65815-2
Koskimäki, J. J., Hokkanen, J., Jaakola, L., Suorsa, M., Tolonen, A., Mattila, S., et al. (2009). Flavonoid biosynthesis and degradation play a role in early defence responses of bilberry (Vaccinium myrtillus) against biotic stress. Eur. J. Plant Pathol. 125, 629–640. doi:10.1007/s10658-009-9511-6
Krcatović, E., Rusak, G., Bezić, N., and Krajačić, M. (2008). Inhibition of tobacco mosaic virus infection by quercetin and vitexin. Acta Virol. 52, 119–124.
Kumar, Y., Dholakia, B. B., Panigrahi, P., Kadoo, N. Y., Giri, A. P., and Gupta, V. S. (2015). Metabolic profiling of chickpea-fusarium interaction identifies differential modulation of disease resistance pathways. Phytochemistry 116, 120–129. doi:10.1016/j.phytochem.2015.04.001
Kumaraswamy, K. G., Kushalappa, A. C., Choo, T. M., Dion, Y., and Rioux, S. (2011). Mass spectrometry based metabolomics to identify potential biomarkers for resistance in barley against fusarium head blight (Fusarium graminearum). J. Chem. Ecol. 37, 846–856. doi:10.1007/s10886-011-9989-1
Kuo, C. L., Agrawal, D. C., Chang, H. C., Chiu, Y. T., Huang, C. P., Chen, Y. L., et al. (2015). In vitro culture and production of syringin and rutin in Saussurea involucrata (kar. Et kir.) – An endangered medicinal plant. Bot. Stud. 56 (1), 12. doi:10.1186/s40529-015-0092-8
Lacroix, D., Prado, S., Kamoga, D., Kasenene, J., and Bodo, B. (2011). Structure and in vitro antiparasitic activity of constituents of Citropsis articulata root bark. J. Nat. Prod. 74, 2286–2289. doi:10.1021/np2004825
Le, J., Ji, H., Zhou, X., Wei, X., Chen, Y., Fu, Y., et al. (2021). pharmacology, toxicology, and metabolism of sennoside a, a medicinal plant-derived natural compound. Front. Pharmacol. 12, 714586. doi:10.3389/fphar.2021.714586
Le, Q. U., Joshi, R. K., Lay, H. L., and Wu, M. C. (2018). Agrimonia pilosa ledeb: Phytochemistry, ethnopharmacology, pharmacology of an important traditional herbal medicine. J. Pharmacol. Phytochem. 7, 3202–3211.
Liebenberg, M. M., Liebenberg, A. J., and Pretorius, Z. (2005). Breeding for rust resistance increases dry bean productivity in South Africa. South Afr. J. Plant Soil 22, 129–139. doi:10.1080/02571862.2005.10634695
Liebenberg, M. M., and Pretorius, Z. A. (2004). Inheritance of resistance to Uromyces appendiculatus in the South African dry bean cultivar kranskop. South Afr. J. Plant Soil 21, 245–250. doi:10.1080/02571862.2004.10635057
Lindgren, D. T., Eskridge, K. M., Steadman, J. R., and Schaaf, D. M. (1995). A model for dry bean yield loss due to rust. HortTechnology 5, 35–37. doi:10.21273/horttech.5.1.35
Lindsey, B. E., Rivero, L., Calhoun, C. S., Grotewold, E., and Brkljacic, J. (2017). Standardized method for high-throughput sterilization of arabidopsis seeds. JoVE 128, e56587. doi:10.3791/56587
Link, T. I. (2020). Testing reference genes for transcript profiling in Uromyces appendiculatus during urediospore infection of common bean. Plos One 15, e0237273. doi:10.1371/journal.pone.0237273
Liu, C., Guo, H., Liu, H., Yu, J., Li, S., Zhu, T., et al. (2022). Differential metabolomics reveals pathogenesis of pestalotiopsis kenyana causing leaf spot disease of Zanthoxylum schinifolium. J. Fungi 8, 1208. doi:10.3390/jof8111208
Liu, W., Cheng, X., Kang, R., Wang, Y., Guo, X., Jing, W., et al. (2021). Systematic characterization and identification of saikosaponins in extracts from Bupleurum marginatum var. stenophyllum using UPLC-PDA-Q/TOF-MS. Front. Chem. 9, 747987. doi:10.3389/fchem.2021.747987
López-Gresa, M. P., Maltese, F., Bellés, J. M., Conejero, V., Kim, H. K., Choi, Y. H., et al. (2010). Metabolic response of tomato leaves upon different plant–pathogen interactions. Phytochem. Anal. Int. J. Plant Chem. Biochem. Tech. 21 (1), 89–94. doi:10.1002/pca.1179
Macel, M. (2011). Attract and deter: A dual role for pyrrolizidine alkaloids in plant–insect interactions. Phytochem. Rev. 10, 75–82. doi:10.1007/s11101-010-9181-1
Makhumbila, P., Rauwane, M., Muedi, H., and Figlan, S. (2022). Metabolome profiling: A breeding prediction tool for legume performance under biotic stress conditions. Plants 11, 1756. doi:10.3390/plants11131756
Manoj, B. S., Gupta, M., Jeelani, M. I., and Gupta, S. (2022). Metabolic footprints of chitosan primed red kidney bean under restricted irrigation. Macromolecules 208, 367–380. doi:10.1016/j.ijbiomac.2022.03.097
Moloto, M. R., Phan, A. D. T., Shai, J. L., Sultanbawa, Y., and Sivakumar, D. (2020). Comparison of phenolic compounds, carotenoids, amino acid composition, in vitro antioxidant and anti-diabetic activities in the leaves of seven cowpea (Vigna unguiculata) cultivars. Foods 9, 1285. doi:10.3390/foods9091285
Montejo Domínguez, L. D. M. A., McClean, P. E., Steadman, J., McCoy, S., Markell, S., and Osorno, J. M. (2022). Bean rust resistance in the Guatemalan climbing bean germplasm collection. Legume Scie 4, e149. doi:10.1002/leg3.149
Mukankusi, C., Raatz, B., Nkalubo, S., Berhanu, F., Binagwa, P., Kilango, M., et al. (2019). Genomics, genetics and breeding of common bean in Africa: A review of tropical legume project. Plant Breed. 138, 401–414. doi:10.1111/pbr.12573
Munakata, K. (1975). Insect antifeeding substances in plant leaves. Pure Appl. Chem. 42, 57–66. doi:10.1016/B978-0-408-70708-4.50008-9
Nagamatsu, A., Masuta, C., Senda, M., Matsuura, H., Kasai, A., Hong, J. S., et al. (2007). Functional analysis of soybean genes involved in flavonoid biosynthesis by virus-induced gene silencing. Plant Biotech. J. 5, 778–790. doi:10.1111/j.1467-7652.2007.00288.x
Obongoya, B. O., Wagai, S. O., and Odhiambo, G. (2010). Phytotoxic effect of selected crude plant extracts on soil-borne fungi of common bean. Afr. Crop Scie. J. 18. doi:10.4314/acsj.v18i1.54189
Odogwu, B. A., Nkalubo, S. T., Mukankusi, C., Odong, T., Awale, H. E., Rubaihayo, P., et al. (2017). Phenotypic and genotypic screening for rust resistance in common bean germplasm in Uganda. Euphytica 213, 49–14. doi:10.1007/s10681-016-1795-y
Ohiagu, F. O., Chikezie, P. C., and Chikezie, C. M. (2021). Toxicological significance of bioactive compounds of plant origin. Pharmacogn. Commun. 11, 67–77. doi:10.5530/pc.2021.2.15
Omara, R. I., Kamel, S. M., El-Ganainy, S. M., Arafa, R. A., Mostafa, Y. S., Alamri, S. A., et al. (2022). Host resistance to Uromyces appendiculatus in common bean genotypes. Plants 11, 628. doi:10.3390/plants11050628
Pluskal, T., Castillo, S., Villar-Briones, A., and Orešič, M. (2010). MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinforma. 11, 395. doi:10.1186/1471-2105-11-395
Puthoff, D. P., Neelam, A., Ehrenfried, M. L., Scheffler, B. E., Ballard, L., Song, Q., et al. (2008). Analysis of expressed sequence tags from Uromyces appendiculatus hyphae and haustoria and their comparison to sequences from other rust fungi. Phytopathology 98, 1126–1135. doi:10.1094/PHYTO-98-10-1126
Quan, L. J., Zhang, B., Shi, W. W., and Li, H. Y. (2008). Hydrogen peroxide in plants: A versatile molecule of the reactive oxygen species network. J. Integr. Plant Biol. 50, 2–18. doi:10.1111/j.1744-7909.2007.00599.x
Ramabulana, A. T., Petras, D., Madala, N. E., and Tugizimana, F. (2021). Metabolomics and molecular networking to characterize the chemical space of four momordica plant species. Metabolites 11, 763. doi:10.3390/metabo11110763
Ramalingam, A., Kudapa, H., Pazhamala, L. T., Weckwerth, W., and Varshney, R. K. (2015). Proteomics and metabolomics: Two emerging areas for legume improvement. Front. Plant Scie. 6, 1116. doi:10.3389/fpls.2015.01116
Raza, A. (2020). Metabolomics: A systems biology approach for enhancing heat stress tolerance in plants. Plant Cell Rep. 29, 741–763. doi:10.1007/s00299-020-02635-8
Robison, F. M., Turner, M. F., Jahn, C. E., Schwartz, H. F., Prenni, J. E., Brick, M. A., et al. (2018). Common bean varieties demonstrate differential physiological and metabolic responses to the pathogenic fungus Sclerotinia sclerotiorum: Bean-sclerotinia physiology and metabolism. Plant Cell Environ. 41, 2141–2154. doi:10.1111/pce.13176
Rosales, M. A., Ocampo, E., Rodríguez-Valentín, R., Olvera-Carrillo, Y., Acosta-Gallegos, J., and Covarrubias, A. A. (2012). Physiological analysis of common bean (Phaseolus vulgaris L.) cultivars uncovers characteristics related to terminal drought resistance. Plant Physiol. biochem. 56, 24–34. doi:10.1016/j.plaphy.2012.04.007
Rubiales, D., Castillejo, M. A., Madrid, E., Barilli, E., and Rispail, N. (2011). Legume breeding for rust resistance: Lessons to learn from the model Medicago truncatula. Euphytica 180, 89–98. doi:10.1007/s10681-011-0367-4
Sadohara, R. (2020). Quality characteristics of bean paste as a confectionery ingredient and recent breeding efforts of common beans in Japan. J. Scie. Food Agric. 100, 10–15. doi:10.1002/jsfa.10013
Saleh, H. M., Hassan, A. A., Mansour, E. H., Fahmy, H. A., and El-Bedawey, A. E. F. A. (2019). Melatonin, phenolics content and antioxidant activity of germinated selected legumes and their fractions. J. Saudi Socie. Agric. Scie. 18, 294–301. doi:10.1016/j.jssas.2017.09.001
Shun-Cheng, R. (2012). Proximate composition and flavonoids content and in vitro antioxidant activity of 10 varieties of legume seeds grown in China. J. Med. Plants Res. 6. doi:10.5897/JMPR11.1408
Singh, G., and Gupta, S. K. (2019). Role of temperature, relative humidity and rainfall in the development of French bean rust (Uromyces appendiculatus). Indian Phytopathol. 72, 271–280. doi:10.1007/s42360-019-00133-w
Singh, P., Arif, Y., Bajguz, A., and Hayat, S. (2021). The role of quercetin in plants. Plant Physio. Biochem. 166, 10–19. doi:10.1016/j.plaphy.2021.05.023
Singh, S. P., and Schwartz, H. F. (2010). Breeding common bean for resistance to diseases: A review. Crop Scie 50, 2199–2223. doi:10.2135/cropsci2009.03.0163
Singla, P., Bhardwaj, R. D., Kaur, S., Kaur, J., and Grewal, S. K. (2020). Metabolic adjustments during compatible interaction between barley genotypes and stripe rust pathogen. Plant physio. biochem. 147, 295–302. doi:10.1016/j.plaphy.2019.12.030
Souza, T. L. P., Faleiro, F. G., Dessaune, S. N., Paula-Junior, T. J. D., Moreira, M. A., and Barros, E. G. D. (2013). Breeding for common bean (Phaseolus vulgaris L.) rust resistance in Brazil. Trop. Plant Path 38, 361–374. doi:10.1590/s1982-56762013005000027
Souza, T. L. P. O., Alzate-Marin, A. L., Faleiro, F. G., and Barros, E. G. (2008). Pathosystem common bean-Uromyces appendiculatus: Host resistance, pathogen specialization, and breeding for rust resistance. Pest Technol. 2, 56–69.
Stein, O., and Granot, D. (2019). An overview of sucrose synthases in plants. Front. Plant Scie. 10, 95. doi:10.3389/fpls.2019.00095
Steinkellner, S., Lendzemo, V., Langer, I., Schweiger, P., Khaosaad, T., Toussaint, J. P., et al. (2007). Flavonoids and strigolactones in root exudates as signals in symbiotic and pathogenic plant-fungus interactions. Molecules 12, 1290–1306. doi:10.3390/12071290
Tashkhodjaev, B., and Sultankhodjaev, M. N. (2009). 8β-acetoxy-14α-benzoyloxy- n -ethyl-3α,10β,13β,15α-tetrahydroxy-1α,6α,16β-trimethoxy-4β-(methoxymethylene)aconitane: Aconifine from aconitum karakolicum rapaics. acta crystallogr. sect. struct. rep. online 65, o1543–o1544. doi:10.1107/s1600536809021436
Tayeh, N., Aubert, G., Pilet-Nayel, M. L., Lejeune-Hénaut, I., Warkentin, T. D., and Burstin, J. (2015). Genomic tools in pea breeding programs: status and perspectives. Front. Plant Scie. 6. doi:10.3389/fpls.2015.01037
Tempesta, M. S., Kriek, G. R., and Bates, R. B. (1982). Uvaricin, a new antitumor agent from Uvaria accuminata (Annonaceae). J. Org. Chem. 47, 3151–3153. doi:10.1021/jo00137a024
Thawabteh, A., Juma, S., Bader, M., Karaman, D., Scrano, L., Bufo, S. A., et al. (2019). The biological activity of natural alkaloids against herbivores, cancerous cells and pathogens. Toxins 11, 656. doi:10.3390/toxins11110656
Thottempudi, V., and Shreeve, J. N. M. (2011). Synthesis and promising properties of a new family of high-density energetic salts of 5-nitro-3-trinitromethyl-1h-1,2,4-triazole and 5,5′-bis(trinitromethyl)-3,3′-azo-1h-1,2,4-triazole. J. Am. Chem. Soc. 133, 19982–19992. doi:10.1021/ja208990z
Toffolatti, S. L., Maddalena, G., Passera, A., Casati, P., Bianco, P. A., and Quaglino, F. (2021). “Role of terpenes in plant defense to biotic stress,” in Biocontrol agents and secondary metabolites (Sawston, United Kingdom: Woodhead Publishing), 401–417. doi:10.1016/b978-0-12-822919-4.00016-8
Tugizimana, F., Piater, L., and Dubery, I. (2013). Plant metabolomics: A new frontier in phytochemical analysis. South Afr. J. scie. 109, 11. doi:10.1590/sajs.2013/20120005
Turetschek, R., Desalegn, G., Epple, T., Kaul, H. P., and Wienkoop, S. (2017). Key metabolic traits of pisum sativum maintain cell vitality during didymella pinodes infection: Cultivar resistance and the microsymbionts influence. J. Proteomics 169, 189–201. doi:10.1016/j.jprot.2017.03.001
Vardavas, C. I., Majchrzak, D., Wagner, K. H., Elmadfa, I., and Kafatos, A. (2006). The antioxidant and phylloquinone content of wildly grown greens in crete. Food Chem. 99, 813–821. doi:10.1016/j.foodchem.2005.08.057
Vasas, A., and Hohmann, J. (2014). Euphorbia diterpenes: Isolation, structure, biological activity and synthesis (2008 -2012). Chem. Rev. 114, 8579–8612. doi:10.1021/cr400541j
Vega, M. P., Santalla, M., and Marsolais, F. (2017). Prospects: The importance of common bean as a model crop. Common Bean Geno, 289–295. doi:10.1007/978-3-319-63526-2_13
Wahl, R., Wippel, K., Goos, S., Kämper, J., and Sauer, N. (2010). A novel high-affinity sucrose transporter is required for virulence of the plant pathogen Ustilago maydis. Plos Biol. 8, e1000303. doi:10.1371/journal.pbio.1000303
Wang, M., Carver, J. J., Phelan, V. V., Sanchez, L. M., Garg, N., Peng, Y., et al. (2016). Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotech. 34, 828–837. doi:10.1038/nbt.3597
Wang, Y. Q., Liu, Q. S., Du, L. X., Hallerman, E. M., and Li, Y. H. (2020). Transcriptomic and metabolomic responses of rice plants to Cnaphalocrocis medinalis caterpillar infestation. Insects 11, 705. doi:10.3390/insects11100705
Watanabe, S., Matsumoto, M., Hakomori, Y., Takagi, H., Shimada, H., and Sakamoto, A. (2014). The purine metabolite allantoin enhances abiotic stress tolerance through synergistic activation of abscisic acid metabolism. Plant Cell & Environ 37, 1022–1036. doi:10.1111/pce.12218
Keywords: Uromyces appendiculatus, rust, Phaseolus vulgaris, common bean, metabolomics, LC-MS
Citation: Makhumbila P, Rauwane ME, Muedi HH, Madala NE and Figlan S (2023) Metabolome profile variations in common bean (Phaseolus vulgaris L.) resistant and susceptible genotypes incited by rust (Uromyces appendiculatus). Front. Genet. 14:1141201. doi: 10.3389/fgene.2023.1141201
Received: 10 January 2023; Accepted: 10 February 2023;
Published: 16 March 2023.
Edited by:
Maryke T. Labuschagne, University of the Free State, South AfricaReviewed by:
Arun Kumar, Institute of Himalayan Bioresource Technology (CSIR), IndiaRashmi Yadav, National Bureau of Plant Genetic Resources (ICAR), India
Copyright © 2023 Makhumbila, Rauwane, Muedi, Madala and Figlan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Penny Makhumbila, 57994463@mylife.unisa.ac.za
 Penny Makhumbila
Penny Makhumbila Molemi E. Rauwane1,2
Molemi E. Rauwane1,2 Hangwani H. Muedi
Hangwani H. Muedi Ntakadzeni E. Madala
Ntakadzeni E. Madala Sandiswa Figlan
Sandiswa Figlan