- 1Department of Herbal Crop Research, National Institute of Horticultural and Herbal Science, Rural Development Administration, Eumseong, South Korea
- 2Division of Botany, Honam National Institute of Biological Resources, Mokpo, South Korea
- 3Department of Biological Sciences, Chungnam National University, Daejeon, South Korea
- 4Department of Biology, Chungbuk National University, Cheongju, South Korea
- 5Department of Industrial Plant Science and Technology, Chungbuk National University, Cheongju, South Korea
- 6Department of Horticulture, Chungbuk National University, Cheongju, South Korea
- 7Department of Crop Science, Chungbuk National University, Cheongju, South Korea
Background: Vicia bungei is an economically important forage crop in South Korea and China. Although detailed genetic and genomic data can improve population genetic studies, conservation efforts, and improved breeding of crops, few such data are available for Vicia species in general and none at all for V. bungei. Therefore, the main objectives of this study were to sequence, assemble, and annotate V. bungei chloroplast genome and to identify simple sequence repeats (SSRs) as polymorphic genetic markers.
Results: The whole-genome sequence of V. bungei was generated using an Illumina MiSeq platform. De novo assembly of complete chloroplast genome sequences was performed for the low-coverage sequence using CLC Genome Assembler with a 200–600-bp overlap size. Vicia bungei chloroplast genome was 130,796-bp long. The genome lacked an inverted repeat unit and thus resembled those of species in the inverted repeat-lacking clade within Fabaceae. Genome annotation using Dual OrganellarGenoMe Annotator (DOGMA) identified 107 genes, comprising 75 protein-coding, 28 transfer RNA, and 4 ribosomal RNA genes. In total, 432 SSRs were detected in V. bungei chloroplast genome, including 64 mononucleotides, 14 dinucleotides, 5 trinucleotides, 4 tetranucleotides, 233 pentanucleotides, 90 hexanucleotides, and 14 complex repeated motifs. These were used to develop 232 novel chloroplast SSR markers, 39 of which were chosen at random to test amplification and genetic diversity in Vicia species (20 accessions from seven species). The unweighted pair group method with arithmetic mean cluster analysis identified seven clusters at the interspecies level and intraspecific differences within clusters.
Conclusion: The complete chloroplast genome sequence of V. bungei was determined. This reference genome should facilitate chloroplast resequencing and future searches for additional genetic markers using population samples. The novel chloroplast genome resources and SSR markers will greatly contribute to the conservation of the genus Vicia and facilitate genetic and evolutionary studies of this genus and of other higher plants.
Introduction
Vicia L. is a genus in the Fabaceae family containing approximately 180–210 species. These species are widely distributed across temperate regions of the northern hemisphere and extend to temperate regions of South America and tropical Africa (Hanelt and Mettin, 1989). Vicia species are used as green manure, cover, forage, and honey crops, making it an economically important genus and a valuable genetic resource (Montemurro et al., 2013). Despite their high economic value, very few genetic and genomic data are available for species of Vicia, other than Vicia villosa (hairy vetch) and Vicia faba (broad bean). Vicia bungei, native to South Korea and China, is phenotypically and ecologically similar to Vicia americana (Endo et al., 2000). Although several chloroplast genomes of different Vicia species have been obtained through next-generation sequencing (NGS) (Cooper et al., 2017; Li et al., 2018; Xin and Yang, 2020), to date, no genetic or genomic studies have been conducted on V. bungei, despite the potentially valuable genetic resources present in wild varieties of this species. In addition, more effective molecular markers are required to support the phylogenetic and population genetic studies underlying the identification, conservation, utilization, and breeding of Vicia species.
The chloroplast genome has long been a focus of research into plant molecular evolution and systematics because of its small size, high copy number, and conservation among species. It has been extensively characterized at the molecular level (Kim et al., 2005). Recent technical advances in NGS technologies mean that the number of completely sequenced chloroplast genomes has increased rapidly, and such sequences play a progressively important role in the identification of molecular markers and in molecular phylogenetic analyses (Kim et al., 2016). Chloroplast genetic markers are potentially more effective indicators of population subdivision and differentiation than are nuclear markers (Schaal et al., 1998; Petit et al., 2005). Chloroplast simple sequence repeats (cpSSRs), generally defined as microsatellites with tandem repeats of 1–6 bp, are valuable resources for assessing genetic and genome diversity, as well as in phylogenetic and systematic evolutionary analyses; cpSSRs have several advantageous characteristics, including haploidy, non-recombination, uniparental inheritance, and low nucleotide substitution rate (Powell et al., 1995). The chloroplast genome can provide unique insight into evolutionary processes as it retains ancient genetic patterns (Provan et al., 2001). Moreover, as the genetic information in angiosperm chloroplasts is inherited maternally, chloroplast markers serve as useful indicators of maternal ancestry (Raveendar et al., 2015).
A new complete chloroplast genome sequence for V. bungei has been generated and compared with sequences of related genera in the Fabaceae family. This comparison enabled the development of cpSSR markers for future population genetics studies, explaining the structure. Our findings help explaining the structure of the complete V. bungei chloroplast genome and reveal evolutionary relationships within the genus Vicia.
Materials and Methods
Plant Material and DNA Extraction
Leaves from 1-year-old V. bungei plants were collected from the Industrial Plant Science and Technology greenhouse at Chungbuk National University (Cheongju, South Korea; 36° 37′ 44.3″ N, 127° 27′ 02.5″ E), immediately snap frozen in liquid nitrogen, and stored at −80°C until analysis. DNA was extracted using automated QIACube system (Qiagen, Hilden, Germany) with a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Qualitative and quantitative assessment of the DNA samples was performed by spectrophotometry (NanoDrop; Thermofisher, Waltham, MA, United States) before their use in the DNA Sequencing.
Sequencing, Assembly, Phylogenetic Relationships, and Comparison of Chloroplast Genome
DNA Sequencing was conducted using the Illumina MiSeq sequencing platform (Illumina, San Diego, CA, United States), and 2.0 Gb of sequence data was generated. Quality trimming and assembly of the reads were achieved using the dnaLCW method (Kim et al., 2015) and CLC Assembly Cell version 4.21 (CLC Inc., Qiagen, Aarhus, Denmark). Phylogenetic relationships were analyzed with the maximum-likelihood method using 61 conserved chloroplast protein sequences from 20 Fabaceae species (downloaded from GenBank; see Supplementary Figure 1) and V. bungei. The analysis was conducted in MEGA7 (Kumar et al., 2016) with 1,000 bootstrap replicates. The complete chloroplast genome of V. bungei was compared with five published chloroplast genomes (Vicia sativa, V. faba, Vicia sepium, Vicia ramuliflora, and Vicia costata) using the mVISTA program (Mayor et al., 2000).
Chloroplast Simple Sequence Repeat Detection and Primer Design
Simple sequence repeat mining was performed using the MIcroSAtellite identification tool (Thiel et al., 2003). The following search parameters were set for identification: mono-, di-, tri-, tetra-, penta-, and hexa-nucleotide motifs with a minimum of ten, five, four, three, two, and two repeats, respectively. The primers for SSR markers were designed using Primer 3.0 software.1 The parameters for designing the primers were set as follows: a primer length of 18–22 bp, with 20 bp set as the optimum value; an optimum annealing temperature of 58°C; and a polymerase chain reaction (PCR) product size of 100–300 bp.
Chloroplast Simple Sequence Repeat Marker Validation and Data Analysis
A total of 39 developed CpSSR markers were randomly selected to assess the genetic diversity of Vicia species (20 accessions from seven species; Supplementary Table 1). The PCR mixture (total volume 40 μl) contained 20 ng genomic DNA, 10 pmol each primer, 2.5 mM MgCl2, 0.25 mM dNTPs, and 0.5 U Taq polymerase (Inclone, Deajeon, South Korea). Polymerase chain reaction amplification was performed under the following conditions: 94°C for 1 min; 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 5 min. The size of PCR products was analyzed using the Fragment Analyzer (Advanced Analytical Technologies Inc., Ankeny, IA, United States), and allele sizes were scored using the PROSize 2.0 (Advanced Analytical Technologies). The number of alleles, the major allele frequency, the expected heterozygosity and polymorphic information content were calculated using the PowerMarker v3.25.2
The expected heterozygosity formula is as follows:
A closely related diversity measure is the polymorphism information content (PIC):
Phylogenetic analysis of Vicia species (20 accessions from seven species) was performed using UPGMA cluster analysis, and unrooted tree construction was based on the CS chord 1967 distance method in PowerMarker v3.25 software.
Results
Genomic Characteristics and Phylogenetic Relationships With Fabaceae
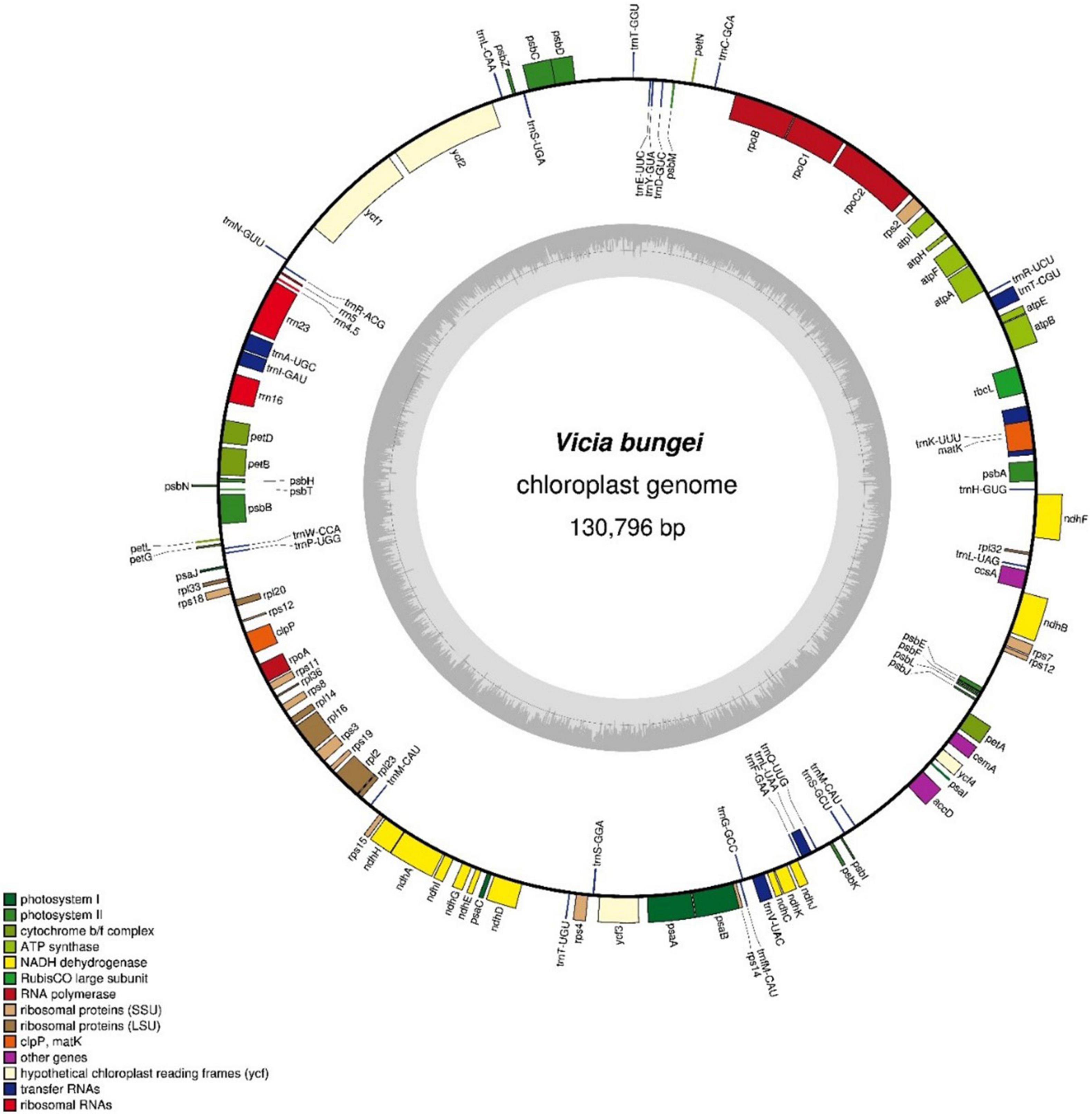
The complete chloroplast genome of V. bungei was 130,796-bp long and lacked an inverted repeat unit (Figure 1). The overall GC content was 34.73%. A total of 107 genes were identified, including 75 protein-coding, 28 transfer RNA, and 4 ribosomal RNA genes (DNA-directed RNA polymerase genes) (Supplementary Table 2). Furthermore, 18 ribosomal subunit genes (ten small subunits and eight large subunits) were detected. Eleven genes, including petB, petD, atpF, ndhA, ndhB, rpl16, rpl2, rps12, rpoC1, clpP, and ycf3, contained one or two introns. Additionally, rps12 was identified as a trans-splicing gene.
A total of 45 chloroplast genes were detected and involved in photosynthesis, and encoded subunits of NADH oxidoreductase (11 genes), subunits of photosystem I (seven genes), subunits of photosystem II (14 genes), subunits of the cytochrome b6/f complex (seven genes), different subunits of ATP synthase (seven genes), and the large chain of ribulose bisphosphate carboxylase (one gene). In addition, five genes were involved in different functions, which two of them remained unknown (Table 1). Vicia bungei chloroplast genome resembled to other Vicia species plastomes in the inverted repeat-lacking clade (IRLC), as it lacked rpl22, rps16, and one intron of clpP (Kim et al., 2016; Li et al., 2018; Xin and Yang, 2020). The complete chloroplast genome sequence, with gene annotations, was submitted to GenBank (accession number MT362055).
Phylogenetic analysis for 21 Fabaceae species, based on 61 conserved plastid protein sequences, showed that V. bungei clustered with two species from the same genus, V. sepium and V. sativa, supported by high bootstrap values in the maximum-likelihood tree (Supplementary Figure 1).
Development of Chloroplast Simple Sequence Repeat Markers
A total of 432 simple sequence repeats (SSRs) were detected, which included 64 mononucleotides, 14 dinucleotides, 5 trinucleotides, 4 tetranucleotides, 233 pentanucleotides, 98 hexanucleotides, and 14 complex repeated motifs, in the chloroplast genome of V. bungei (Supplementary Figure 2). The majority of the identified SSRs were pentanucleotide repeats (53.9%), followed by hexanucleotides repeats (22.7%). A total of 432 potential SSR motifs were identified, 313 (73.45%) of which occurred within the intergenic regions (Supplementary Table 3).
A total of 232 pairs of SSR primers were designed from 432 potential SSR motifs (Supplementary Table 4) that showed potential for marker development; these included mono-, di-, tetra-, penta-, and hexanucleotides, and complex repeated motifs. Pentanucleotides (54.31%) were the most abundant group within the selected SSR markers, followed by hexa- (25%), mono- (14.65%), di- (6%), and tetranucleotides (4%) and complex repeated nucleotide (4%) markers (Supplementary Table 4). The most common motif from the pentanucleotide markers was the GAATT/GAAAT (4.76%), followed by CAAAA/CATAA (3.97%), AAAGA/AATGA (3.97%), and TATAT/TATTT (3.17%) (Supplementary Table 4).
Genetic Diversity in Vicia Species
Of the 232 cpSSR markers described above, 39 were selected randomly, in order to evaluate their amplification potential and to assess genetic diversity in the genus Vicia (20 accessions from seven species). Amplification of all the selected cpSSR markers produced clear fragments; 35 fragments showed polymorphisms, and four were monomorphic. The major allele frequencies within the 39 SSR markers across 20 accessions (Vicia spp.) ranged from 0.20 (VBCP38 and VBCP42) to 1.0 (VBCP41, VBC54, VBC65, and VBC164), with a mean value of 0.62. The number of alleles per marker varied between 1 (VBCP41, VBC54, VBC65, and VBC164) and 16 (VBCP42), with a mean value of 3.92. The expected heterozygosity ranged from 0 (VBCP41, VBC54, VBC65, and VBC164) to 0.92 (VBCP39), with a mean value of 0.48. The polymorphic information content ranged from 0 (VBCP41, VBC54, VBC65, and VBC164) to 0.91 (VBCP39), with a mean value of 0.569 (Table 2). The most polymorphic loci (i.e., those exhibiting the highest diversity) were VBCP39 and VBCP42, based on their relatively large allele number (15 and 16, respectively).
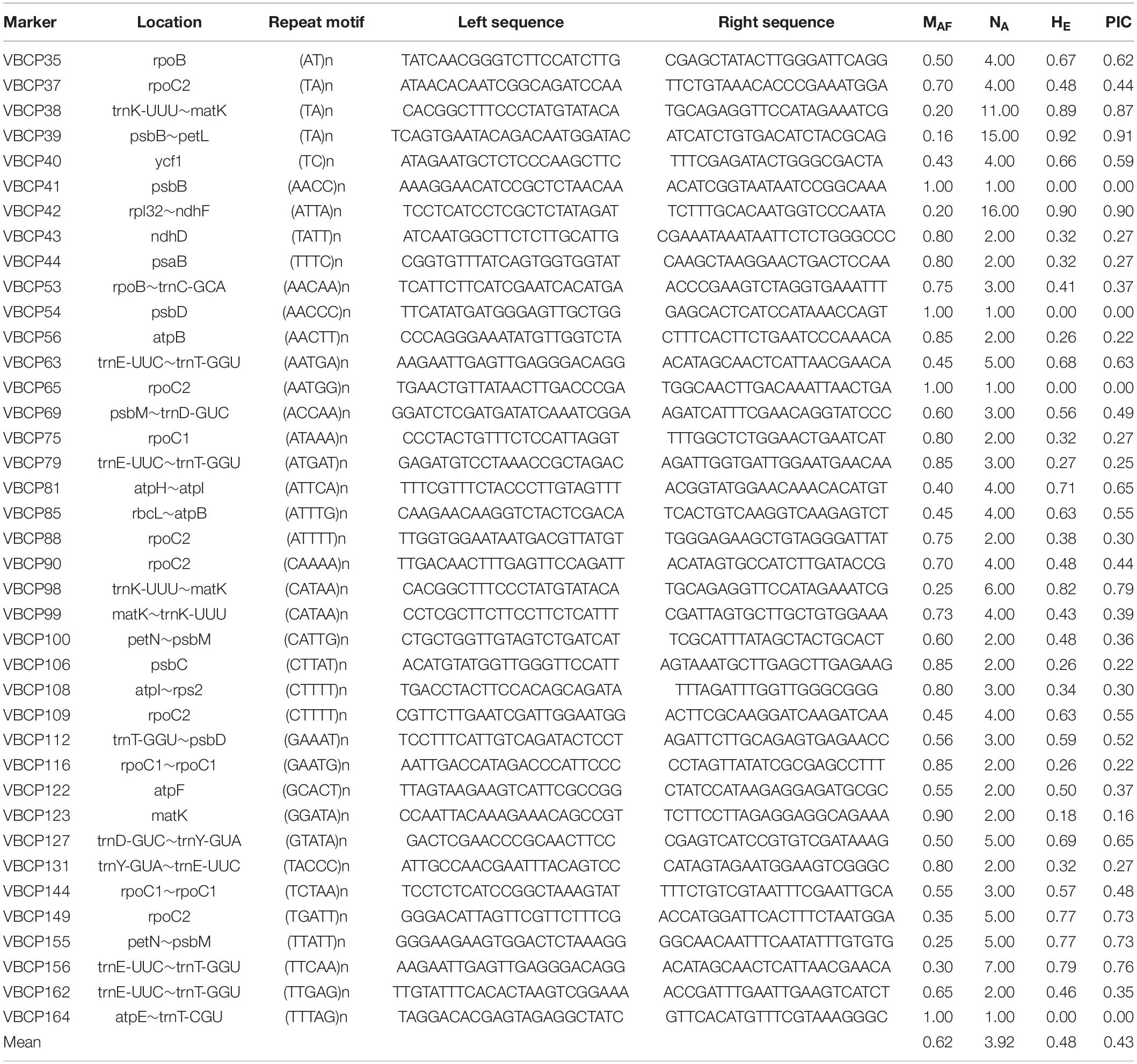
Table 2. Summary of the 39 polymorphic chloroplast simple sequence repeat markers and their genetic diversity statistics across 20 Vicia spp. accessions.
These 39 cpSSR markers were used to further analyze the genetic diversity of the 20 Vicia spp. accessions. The unweighted pair group method with arithmetic mean (UPGMA) cluster phylogenetic analysis clearly distinguished the accessions by genotype and grouped them into seven major clusters that corresponded with the different species (Figure 2). Targeted analysis of cpSSR regions in V. bungei identified a unique chloroplast type for each of the seven species examined (group A: Vicia dasycarpa; group B: Vicia hirsuta; group C: Vicia narbonensis; group D: Vicia angustifolia var. segetilis; group E: V. bungei; group F: Vicia linearifolia; group G: Vicia chosenensis). The UPGMA cluster analysis also revealed intraspecific variation between the 20 Vicia spp. accessions.
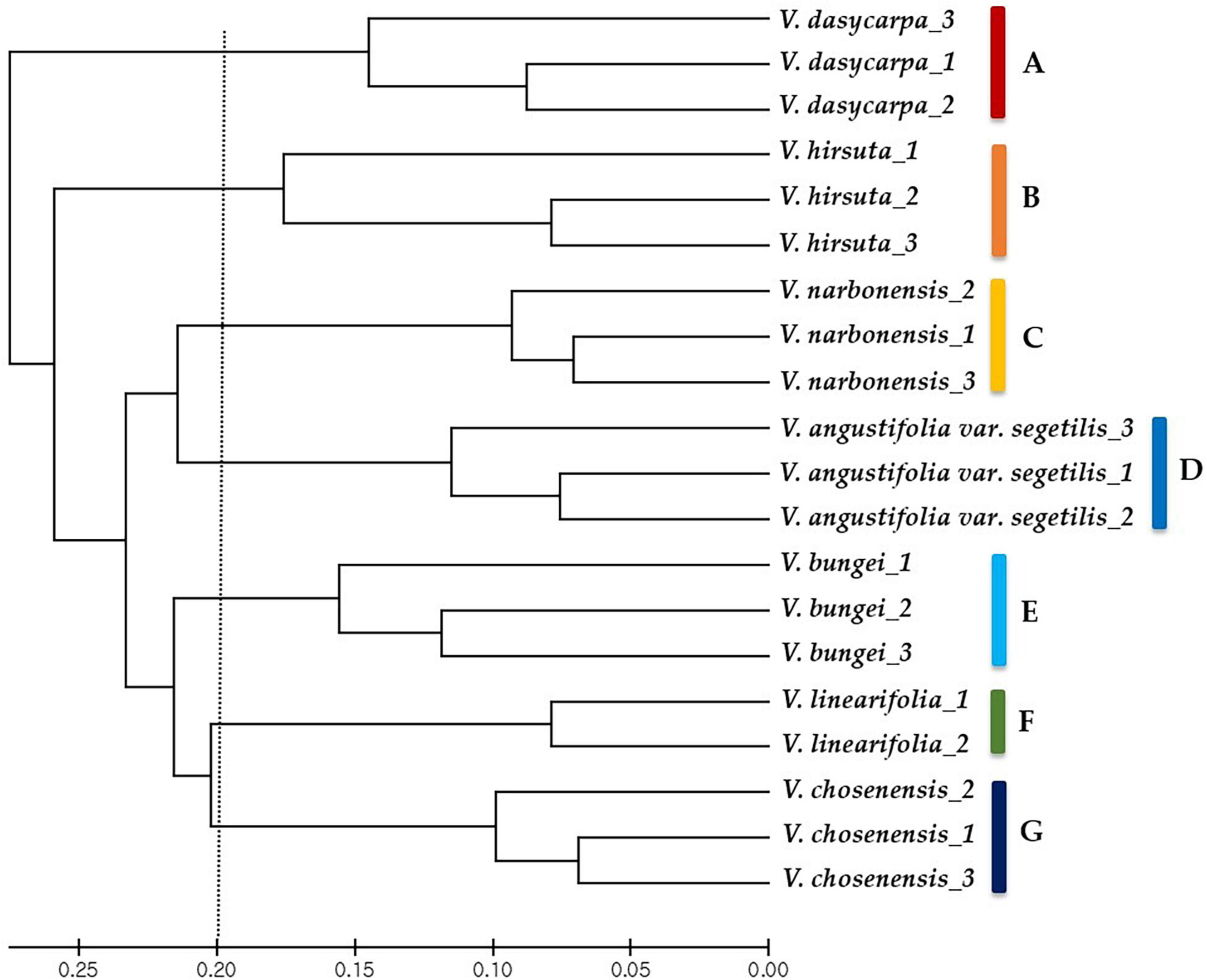
Figure 2. Phylogenetic relationships between 20 Vicia spp. accessions. The phylogenetic tree was constructed with data from 39 chloroplast simple sequence repeat markers using the unweighted pair group method with arithmetic mean method. Letters A–E on the right-hand side represent each plastome group based on the plastome haplotypes.
Discussion
Most chloroplast genomes have a circular structure that contains a large single-copy region, an inverted repeat A region, a small single-copy region, and an inverted repeat B region (Raveendar et al., 2015; Zhu et al., 2016). The chloroplast genomes of some legumes, including V. sepium, have lost one of the two inverted repeats, and these species form the IRLC (Li et al., 2018). The chloroplast genome of V. bungei indicates that this economically important crop is also a member of the IRLC, suggesting that it may be part of the same evolutionary clade as V. sepium and V. sativa, which may share a similar evolutionary history.
A chloroplast gene is rarely lost arbitrarily. Instead, the gene is either transferred to the nuclear genome, or its function is replaced by a nuclear gene (Wang et al., 2018). Recent sequencing and analyses of some IRLC plastomes (Kim et al., 2005; Li et al., 2018; Moghaddam et al., 2022) have revealed important evolutionary patterns in this clade, including loss of the genes rpl22 and rps16, the deletion of one intron of clpP (Jansen et al., 2008; Li et al., 2020), multiple sequence inversions (Schwarz et al., 2015; Williams et al., 2015), and gene transfers to the nucleus (Moghaddam and Kazempour-Osaloo, 2020; Wu et al., 2021). Vicia bungei plastome lacked one intron of clpP, which was consistent with the finding of Jansen et al. (2008) and confirmed the parallel loss of this clpP intron in V. bungei and in members of the papilionoid IRLC. Recently, Williams et al. (2015) demonstrated that the rates of synonymous and non-synonymous mutations are accelerated in the clpP sequence of Acacia, suggesting there may be a functional nuclear-encoded copy of this gene in at least some mimosoid legumes. The mechanism by which rpl22, rps16, and one intron of clpP gene have been lost from V. bungei requires further in-depth research.
Although the structure of plastid genomes is largely conserved across land plants (Wicke et al., 2011), other exceptions to this pattern, beyond these IRLC legumes, include Geraniaceae (Guisinger et al., 2011) and Campanulaceae (Haberle et al., 2008). Some Fabaceae plastomes have been highly rearranged owing to multiple rounds of translocations and/or inversions. As a result, the plastomes of the IRLC have undergone considerable diversification in both gene order and gene/intron content (Wang et al., 2018; Li et al., 2020; Xin and Yang, 2020; Moghaddam et al., 2022).
The plastome of V. bungei most closely resembled those of V. sepium and V. sativa. This finding is consistent with previous research that indicated similar evolutionary evaluation analysis of V. sepium and V. sativa (Li et al., 2020). Plastome differences between some major clades provide valuable information for resolving phylogenetic relationships based on DNA sequence analyses. The study of genomic variation and phylogeny of Fabaceae species provides an increased understanding of general chloroplast genome evolution. In our results of multiple alignments among six Vicia species cp genomes, we observed 11 major variant regions of V. bungei in cp genome (Supplementary Figure 3). However, our research focus on the development of cpSSR markers for Vicia species identification. Thus, these variant regions would be very useful for further evolutionary studies as well as cpSSR marker design.
In other flowering plant chloroplast genomes, the most common SSR motifs are mono-, di-, and trinucleotide repeats (George et al., 2015). Nevertheless, data mining, under the recursive criteria adopted here, revealed that the majority of SSRs present in the V. bungei chloroplast genome sequences contained relatively long penta- (53.9%) or hexanucleotide (22.7%) motifs. In this study, a total of 432 potential SSR were found in the chloroplast genome of V. bungei. The number of repeat motifs was richer in V. bungei chloroplast genome compared with the V. sepium chloroplast genome (Li et al., 2020). These results provide a firm foundation for further investigations of chloroplast genome evolution in Vicia L. and other IRLC legumes such as Medicago and Pisum and species.
Raveendar et al. (2015) used SSR markers to detect genetic diversity in V. sativa and 22 other Vicia species. Li et al. (2018) used chloroplast genome sequences to determine the genetic diversity in V. sepium and 21 closely related Fabaceae species. Han et al. (2021) used barcoding loci (ITS2, matK, and rbcL) as DNA markers to differentiate 19 Vicia taxa. Previous studies of phylogenetic relationships among species of the subgenus Vicia did not detect any intraspecific variation when cDNA SSR, cpSSR, or DNA barcoding markers were applied to representative plants of each species. By contrast, our UPGMA cluster analysis classified seven clusters at the interspecies level and revealed intraspecific differences within clusters. These results suggest that the 39 cpSSR markers developed in V. bungei differentiated efficiently between Vicia species genotypes and also provide estimates of their genetic diversity. These cpSSR markers will thus be useful, not only for authenticating Vicia species but also for providing the baseline data essential for advancing systematic breeding in the field and the development of conservation strategies, as well as for guiding the collection of germplasm.
Conclusion
Complete chloroplast genomes have helped to reveal intraspecies relationships, but also allow to measure divergence within interspecies. Growing genomic resources for Vicia spp. provide tools to extend our knowledge on this critically important forage crop species. To develop cpSSR markers that can be utilized to classify Vicia species and analyze the genetic diversity of related species, potential cpSSR motifs were mined from the chloroplast genome of V. bungei, and finally, 39 cpSSR markers were developed. The UPGMA cluster analysis detected intra-and interspecific variation between 20 accessions with 39 cpSSR markers to distinguish Vicia species. The chloroplast genome and cpSSR markers found in this study would provide useful information for genetic diversity analyses in Vicia species.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
I-HJ and SH performed the experiments, analyzed the data, prepared figures and tables, and wrote the manuscript. DS, HR, TH, YL, DK, Y-SS, and J-WC reviewed drafts of the manuscript. All authors have read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.891783/full#supplementary-material
Footnotes
References
Cooper, J. W., Wilson, M. H., Derks, M. F., Smit, S., Kunert, K. J., Cullis, C., et al. (2017). Enhancing faba bean (Vicia faba L.) genome resources. J. Exp. Bot. 68, 1941–1953. doi: 10.1093/jxb/erx117
Endo, Y., Choi, B. H., and Ohashi, H. (2000). Distinction between Vicia americana and V. bungei (Leguminosae). J. Jpn. Bot. 75, 92–97.
George, B., Bhatt, B. S., Awasthi, M., George, B., and Singh, A. K. (2015). Comparative analysis of microsatellites in chloroplast genomes of lower and higher plants. Curr. Genet. 61, 665–677. doi: 10.1007/s00294-015-0495-9
Guisinger, M. M., Kuehl, J. V., Boore, J. L., and Jansen, R. K. (2011). Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol. Biol. Evol. 28, 583–600. doi: 10.1093/molbev/msq229
Haberle, R. C., Fourcade, H. M., Boore, J. L., and Jansen, R. K. (2008). Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J. Mol. Evol. 66, 350–361. doi: 10.1007/s00239-008-9086-4
Han, S., Sebastin, R., Wang, X., Lee, K. J., Cho, G. T., Hyun, D. Y., et al. (2021). Identification of Vicia species native to South Korea using molecular and morphological characteristics. Front. Plant Sci. 12:608559. doi: 10.3389/fpls.2021.608559
Hanelt, P., and Mettin, D. (1989). Biosystematics of the genus Vicia L. (Leguminosae). Annu. Rev. Ecol. Evol. Syst. 20, 199–223. doi: 10.1146/annurev.es.20.110189.001215
Jansen, R. K., Wojciechowski, M. F., Sanniyasi, E., Lee, S. B., and Daniell, H. (2008). Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae). Mol. Phylogenet. Evol. 48, 1204–1217. doi: 10.1016/j.ympev.2008.06.013
Kim, K., Lee, S. C., Lee, J., Yu, Y., Yang, K., Choi, B. S., et al. (2015). Complete chloroplast and ribosomal sequences for 30 accessions elucidated the evolution of Oryza AA genome species. Sci. Rep. 5:15655. doi: 10.1038/srep15655
Kim, K. J., Choi, K. S., and Jansen, R. K. (2005). Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae). Mol. Biol. Evol. 22, 1783–1792. doi: 10.1093/molbev/msi174
Kim, T. S., Lee, J. R., Raveendar, S., Lee, G. A., Jeon, Y. A., Lee, H. S., et al. (2016). Complete chloroplast genome sequence of Capsicum baccatum var. baccatum. Mol. Breed. 36:110. doi: 10.1007/s11032-016-0532-5
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, C., Zhao, Y., Huang, H., Ding, Y., Hu, Y., and Xu, Z. (2018). The complete chloroplast genome of an inverted-repeat-lacking species, Vicia sepium, and its phylogeny. Mitochondrial DNA B Resour. 3, 137–138. doi: 10.1080/23802359.2018.1431071
Li, C., Zhao, Y., Xu, Z., Yang, G., Peng, J., and Peng, X. (2020). Initial characterization of the chloroplast genome of Vicia sepium, an important wild resource plant, and related inferences about its evolution. Front. Genet. 11:73. doi: 10.3389/fgene.2020.00073
Mayor, C., Brudno, M., Schwartz, J. R., Poliakov, A., Rubin, E. M., Frazer, K. A., et al. (2000). VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16, 1046–1047. doi: 10.1093/bioinformatics/16.11.1046
Moghaddam, M., and Kazempour-Osaloo, S. (2020). Extensive survey of the ycf 4 plastid gene throughout the IRLC legumes: robust evidence of its locus and lineage specific accelerated rate of evolution, pseudogenization and gene loss in the tribe Fabeae. PLoS One 15:e0229846. doi: 10.1371/journal.pone.0229846
Moghaddam, M., Ohta, A., Shimizu, M., Terauchi, R., and Kazempour-Osaloo, S. (2022). The complete chloroplast genome of Onobrychis gaubae (Fabaceae-Papilionoideae): comparative analysis with related IR-lacking clade species. BMC Plant Biol. 22:75. doi: 10.1186/s12870-022-03465-4
Montemurro, F., Fiore, A., Campanelli, G., Tittarelli, F., Ledda, L., and Canali, S. (2013). Organic fertilization, green manure, and vetch mulch to improve organic zucchini yield and quality. Hortscience 48, 1027–1033. doi: 10.21273/HORTSCI.48.8.1027
Petit, R. J., Duminil, J., Fineschi, S., Hampe, A., Salvini, D., and Vendramin, G. G. (2005). Invited review: comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol. Ecol. 14, 689–701. doi: 10.1111/j.1365-294X.2004.02410.x
Powell, W., Morgante, M., McDevitt, R., Vendramin, G. G., and Rafalski, J. A. (1995). Polymorphic simple sequence repeat regions in chloroplast genomes: applications to the population genetics of pines. Proc. Natl. Acad. Sci. U.S.A. 92, 7759–7763. doi: 10.1073/pnas.92.17.7759
Provan, J., Powell, W., and Hollingsworth, P. M. (2001). Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends Ecol. Evol. 16, 142–147. doi: 10.1016/S0169-5347(00)02097-8
Raveendar, S., Lee, G. A., Jeon, Y. A., Lee, Y. J., Lee, J. R., Cho, G. T., et al. (2015). Cross-amplification of Vicia sativa subsp. sativa microsatellites across 22 other Vicia species. Molecules 20, 1543–1550. doi: 10.3390/molecules20011543
Schaal, B. A., Hayworth, D. A., Olsen, K. M., Rauscher, J. T., and Smith, W. A. (1998). Phylogeographic studies in plants: problems and prospects. Mol. Ecol. 7, 465–474. doi: 10.1046/j.1365-294x.1998.00318.x
Schwarz, E. N., Ruhlman, T. A., Sabir, J. S., Hajrah, N. H., Alharbi, N. S., Al-Malki, A. L., et al. (2015). Plastid genome sequences of legumes reveal parallel inversions and multiple losses of rps16 in papilionoids. J. Syst. Evol. 53, 458–468. doi: 10.1111/jse.12179
Thiel, T., Michalek, W., Varshney, R. K., and Graner, A. (2003). Exploiting EST databases for the development and characterization of gene-derived SSR markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 106, 411–422. doi: 10.1007/s00122-002-1031-0
Wang, Y. H., Wicke, S., Wang, H., Jin, J. J., Chen, S. Y., Zhang, S. D., et al. (2018). Plastid genome evolution in the early-diverging legume subfamily Cercidoideae (Fabaceae). Front. Plant Sci. 9:138. doi: 10.3389/fpls.2018.00138
Wicke, S., Schneeweiss, G. M., Depamphilis, C. W., Müller, K. F., and Quandt, D. (2011). The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 76, 273–297. doi: 10.1007/s11103-011-9762-4
Williams, A. V., Boykin, L. M., Howell, K. A., Nevill, P. G., and Small, I. (2015). The complete sequence of the Acacia ligulata chloroplast genome reveals a highly divergent clpP1 gene. PLoS One 10:e0125768. doi: 10.1371/journal.pone.0125768
Wu, S., Chen, J., Li, Y., Liu, A., Li, A., Yin, M., et al. (2021). Extensive genomic rearrangements mediated by repetitive sequences in plastomes of Medicago and its relatives. BMC Plant Biol. 21:421. doi: 10.1186/s12870-021-03202-3
Xin, C., and Yang, Q. (2020). The first complete chloroplast genome sequence of Vicia ramuliflora (Fabaceae). Mitochondrial DNA B Resour. 5, 410–411. doi: 10.1080/23802359.2019.1705196
Keywords: chloroplast genome, inverted repeat-lacking clade, phylogeny, SSR marker, Vicia bungei
Citation: Jo I-H, Han S, Shim D, Ryu H, Hyun TK, Lee Y, Kim D, So Y-S and Chung J-W (2022) Complete Chloroplast Genome of the Inverted Repeat-Lacking Species Vicia bungei and Development of Polymorphic Simple Sequence Repeat Markers. Front. Plant Sci. 13:891783. doi: 10.3389/fpls.2022.891783
Received: 08 March 2022; Accepted: 26 April 2022;
Published: 16 May 2022.
Edited by:
Aliki Xanthopoulou, Hellenic Agricultural Organization (HAO), GreeceReviewed by:
Qingguo Ma, Chinese Academy of Forestry, ChinaGeorgios Tsoktouridis, Hellenic Agricultural Organization Dimitra, Thessaloniki, Greece
Copyright © 2022 Jo, Han, Shim, Ryu, Hyun, Lee, Kim, So and Chung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yoon-Sup So, yoonsupso@chungbuk.ac.kr; Jong-Wook Chung, jwchung73@chungbuk.ac.kr
†These authors have contributed equally to this work and share first authorship
 Ick-Hyun Jo
Ick-Hyun Jo Seahee Han
Seahee Han Donghwan Shim
Donghwan Shim Hojin Ryu4
Hojin Ryu4 Jong-Wook Chung
Jong-Wook Chung