. Introduction
The lichen genus Parmelia (L.) Ach. belongs to the family Parmeliaceae and comprises foliose species with elongated or effigurate pseudocyphellae on the upper surface of the thallus (located marginal or/and laminal), and simple, branched to squarrose rhizines on the lower side (Hale, 1987; Thell et al., 2011). Atranorin is always present in the upper thallus cortex and depsides, depsidones, fatty acids, or, less frequently, dibenzofurans in the medulla (Hale, 1987; Ossowska et al., 2018; Thell et al., 2008). To date, about 41 Parmelia species have been described (Crespo et al., 2020; Divakar et al., 2015), and their identification is based on morphological, chemical, and ecological data, often supported by molecular evidence (Corsie et al., 2019). Such a multifaceted approach has recently been recommended as due to the plasticity of diagnostic features in Parmelia, the determination of specimens based on solely the traditional taxonomic methods can lead to some identification errors (e.g. Castellani et al., 2021; Corsie et al., 2019; Haugan & Timdal, 2019; Ossowska et al., 2018, 2019; Tsurykau et al., 2019). For example, the presence of pruina has been reported as a diagnostic feature in isidiate Parmelia species (Feuerer & Thell, 2002; Molina et al., 2004), but it has no diagnostic value as sampled studies show that individual specimens can vary in the pruinosity degree (Corsie et al., 2019; Ossowska et al., 2018). In the group of Parmelia species without vegetative propagule, the presence of lobaric acid was reported as a diagnostic (Hale, 1987; Thell et al., 2008), however, it has been shown that this secondary metabolite can be present or absent in different specimens of the same species (Ossowska et al., 2019). It proved that the identification of Parmelia on the basis of morphological and chemical characteristics alone cannot be satisfactory. In addition, the incorporation of molecular methods in the taxonomy of the genus Parmelia has contributed to the description of cryptic taxa, such as P. encryptata A. Crespo, Divakar & M. C. Molina (Molina et al., 2011; Ossowska et al., 2021) and near-cryptic, like P. rojoi A. Crespo, V. J. Rico & Divakar, within the genus (Crespo et al., 2020). As a consequence of the widespread application of the integrative taxonomy concept, the knowledge of the distribution of Parmelia species should also be updated.
Here, we report new localities of three Parmelia species, whose identification is supported by molecular data. These are the first localities of P. ernstiae Feuerer & A. Thell from Madeira, three new records of P. encryptata from Poland, and three new records of P. sulcata Taylor from Chile, the last two species being rarely reported in these countries. We also discuss the diagnostic features of these species and their differences with other similar taxa, together with notes on their distribution.
. Materials and methods
Taxon sampling. Fresh material (a total of 50 specimens) for this study was collected during various lichenological fieldwork in Chile in 2023, Madeira in 2022, and Poland in 2017 and 2023 by U. Schiefelbein, R. Szymczyk, and M. Kukwa, respectively. All samples are deposited in the UGDA herbarium. Morphology was examined under a stereomicroscope. Lichen substances were identified with thin-layer chromatography methods (TLC) in solvents A and C according to methods summarized by Orange et al. (2001). The distribution map was prepared using QGIS 3.30.1 software.
DNA extraction, amplification, sequencing and haplotype network. DNA was extracted, amplified, and purified using the instructions presented in previous works (Ossowska et al., 2018, 2019). Sequencing was performed in Macrogen sequencing system (http://www.macrogen.com). The newly obtained sequences were deposited in GenBank (http://www.ncbi.nlm.nih.gov/genbank) and their accession numbers are listed in Table 1. The newly generated nucITS rDNA sequences were compared using BLAST search (Altschul et al., 1990).
Table 1
List of sequences used in haplotype network. Newly obtained sequences are in bold.
For haplotype network analysis, we downloaded all sequences of Parmelia encryptata, P. ernstiae, and P. sulcata (Table 1) from GenBank. All sequences were aligned in Seaview (Galtier et al., 1996; Gouy et al., 2010), and the terminal ends were cut. The alignment of P. encryptata consisted of 20 sequences, P. ernstiae of 123 sequences, and P. sulcata of 194 sequences. The TCS network (Clement et al., 2002) was created using PopArt software (http://popart.otago.ac.nz) and modified in Inkscape (http://inkscape.org).
. Results and discussion
Parmelia encryptata A. Crespo, Divakar & M. C. Molina
Parmelia encryptata is a cryptic species, morphologically and chemically identical to P. sulcata, and is characterized by sublinear lobes (Figure 1A), with marginal and laminal, elongated pseudocyphellae and soralia on the upper surface (Figure 1A, Figure 1B), the presence of salazinic acid in the medulla and black, simple to squarrose rhizines on the lower surface (Figure 1C, Figure 1D) (Molina et al., 2011). In the new specimens, we observed a predominance of simple rhizines, with squarrose rhizines being grouped only in the central part of the thalli (Figure 1C). This trait has been suggested by Ossowska et al. (2021) as a potential diagnostic feature of P. encryptata. However, the number of studied specimens of this species is still too small to unequivocally confirm its usefulness. Furthermore, very few specimens of P. sulcata (e.g., samples from Chile cited below) may show similar characteristics in the rhizines placement to studied samples of P. encryptata; nevertheless the examination of this character can help to select samples for molecular study to confirm their identification.
Figure 1
Morphology of Parmelia encryptata. (A) lobes with marginal and laminal, elongated pseudocyphellae (UGDA L-61390); (B) lobes with soralia (UGDA L-24009); (C) rhizines (UGDA L-24009); (D) sorediate lobes with rhizines visible at the edges (UGDA L-61390). Scale bars: 1 mm.

The nucITS rDNA sequences were obtained from three specimens (Table 1) and BLAST search shows 99% and 100% similarity to the sequence of P. encryptata from Switzerland (MN654571). This sequence was originally deposited by Mark et al. (2020) as P. sulcata, but later corrected to P. encryptata by Ossowska et al. (2021). Furthermore, within the new sequences, we observed six nucleotides that Molina et al. (2011) and Ossowska et al. (2021) identified as diagnostic to distinguish P. encryptata from P. sulcata.
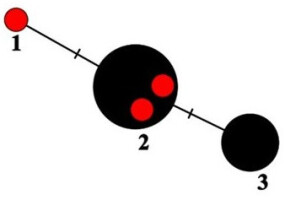
In the haplotype network (Figure 2), the sequences of P. encryptata are represented by three haplotypes (see also Table 1). The two specimens from Poland (UGDA L-24009 and 24214), like the specimens from Białowieża Forest cited by Ossowska et al. (2021), share the same haplotype (no. 2 in Figure 2) with the specimens from Ireland, Spain and Switzerland. The specimen from Borecka Forest (UGDA L-61390, haplotype no. 1) differs from haplotype no. 2 in one site and represents a new haplotype of the species (Figure 2). The haplotype no. 3 is represented by a group of P. encryptata specimens from Spain (Molina et al., 2011), and a single sequence (AF159947, see Table 1) obtained by Arup and Grube (2000), for which there is no information on collection site (therefore the record is not presented in Figure 3). The sequences deposited as P. sulcata in GenBank (OQ717535) obtained from a specimen from the Czech Republic share six nucleotides characteristic of P. encryptata. This sequence also shows similarity with P. encryptata sequences in BLAST search. Unfortunately, there are no geographical coordinates for this specimen in the GenBank database. Therefore, it is not presented in Figure 3.
Figure 2
Haplotype network showing relationships among ITS haplotypes of Parmelia encryptata. The new sequences from Poland are marked as red dots.
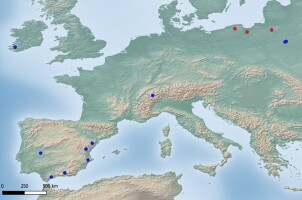
The species is only known in Europe from Ireland, Poland, Spain, Switzerland (Ossowska et al., 2021) (Figure 3), and the Czech Republic (see above). In Poland, it was previously reported only from two localities in the Białowieża Forest (Ossowska et al., 2021). Taking into account all confirmed localities of this taxon, Ossowska et al. (2021) suggested that it prefers large forest ecosystems. However, new record of the species from the cemetery indicate that P. encryptata may also be present in other habitats as well.
Figure 3
Distribution map of Parmelia encryptata. Blue dots – localities according to literature data, red dots – new localities in Poland (base map provided by Natural Earth, www.naturalearthdata.com).
Specimens examined: Poland, (1) Pojezierze Ełckie, Puszcza Borecka Forest, 54.11662°N, 22.07067°E, ATPOL grid square Bf 03, deciduous forest dominated by oaks and hornbeam, on Tilia cordata, 18 March 2023, leg. R. Szymczyk, det. E. A. Ossowska (UGDA L-61390; GenBank OR509530); (2) Pojezierze Dzierzgońsko-Morąskie, Postolin, 53.873889°N, 19.052222°E, ATPOL grid square Bd 43, cemetery with free standing trees, on Tilia cordata, 15 Apr. 2017, leg. M. Kukwa 19243, det. E. A. Ossowska (UGDA L-24009; GenBank OR509531); (3) Wysoczyzna Polanowska, Skotawskie Łąki nature reserve, by unnamed lake (N of Lipieniec lake), 54.261944°N, 17.553056°E, ATPOL grid square Ac 93, beech forest, on Fagus sylvatica, 27 June 2017, leg. M. Kukwa 19830, det. E. A. Ossowska (UGDA L-24214; GenBank OR509532).
Parmelia ernstiae Feuerer & A. Thell
Parmelia ernstiae was previously separated from other isidiate species mainly on the basis of the presence of pruina, placement of isidia and the production of fatty acids and lobaric acid (Feuerer & Thell, 2002; Molina et al., 2004; Ossowska, 2021; Ossowska et al., 2018). However, according to Corsie et al. (2019), P. ernstiae is a near-cryptic species, very similar in morphology and secondary chemistry to P. saxatilis and P. serrana A. Crespo, M. C. Molina & D. Hawksw., and their correct differentiation, therefore, requires the use of a combination of molecular and phenotypic characters. Variation in morphological characters was also observed in specimens from Madeira. In specimens UGDA L-61375 and 61386, we found a set of features that Corsie et al. (2019) and Ossowska et al. (2018) described as most common in P. ernstiae – a short, broad lobes with laminal isidia, which are present predominantly in central parts of thalli (Figure 4C). On the other hand, in specimens UGDA L-61382 and 61384 the lobes were longer and broader, and sublinear (Figure 4A, Figure 4B) as in P. saxatilis. In addition, the isidia were laminal and marginal, as in P. serrana (Figure 4A). The common features for all specimens were non-overlapping lobes very often with lobules, laminal and marginal, linear pseudocyphellae (Figure 4A–D), and simple to furcate rhizines (Figure 4D). The shape of lobes and distribution of isidia are characteristics that may vary between samples, as shown by Corsie et al. (2019). All specimens produce lobaric and fatty acids as reported in previous works, but specimens without these substances are also known from the species (Corsie et al., 2019; Ossowska et al., 2018).
Figure 4
Morphology of Parmelia ernstiae specimens from Madera. (A) and (B) sublinear lobes with marginal and laminal isidia (UGDA L-61382); (B) lobes with marginal and laminal pseudocyphellae (UGDA L-61382); (C) short and broad lobes (UGDA L-61384); (D) lobes with lobules (UGDA L-61384). Scale bars: 1 mm.

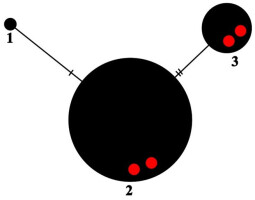
The nucITS rDNA sequences were obtained from four specimens (Table 1) and show 97% (UGDA L-61375 & 61384) and 100% (UGDA L-61386 & 61382) of similarity to the sequences of P. ernstiae deposited in GenBank (OQ717996 and MK567162). In the haplotype network, three haplotypes of P. ernstiae were found (Figure 5). The samples from Madeira represent two haplotypes (no. 2 and 3), which differ from each other by two sites (Figure 5). The haplotype no. 2 seems to be very common, as it is represented by more than one hundred P. ernstiae sequences. Most of these were obtained from specimens collected in Great Britain (Corsie et al., 2019), but nevertheless, this haplotype was observed in specimens collected from fifteen countries (Table 1). The haplotype no. 3, in addition to two samples from Madeira, is also represented by sequences from the Czech Republic, Germany, Poland, Russia, Spain, and Turkey (see also Table 1).
Figure 5
Haplotype network showing relationships among ITS haplotypes of Parmelia ernstiae. The new sequences from Madeira are marked as red dots.
Parmelia ernstiae is widely distributed in Europe (Castellani et al., 2021; Corsie et al., 2019; Feuerer & Thell, 2002; Hawksworth et al., 2008, 2011; Kukwa et al., 2012; Tsurykau et al., 2019). It is worth noting that not all records are supported by phylogenetic evidence. Outside Europe, the species has been reported from Africa (Algeria and the Canary Islands), as well as from the Asian part of Russia (Hawksworth et al., 2008, 2011). Here, we present the first records of P. ernstiae from Madeira (Portugal).
Specimens examined: Portugal, Madeira, (1) Near the road to Abrigo do Poiso village, 32.71917°N, 16.88806°W, forest with Picea sp., on the bark of Picea sp., 30 May 2022, leg. R. Szymczyk, det. E. A. Ossowska (UGDA L-61382 & 61384; GenBank OR509534 & OR509535); (2) Riberio Frio village, edge of deciduous forest, 32.738889°N, 16.886944°W, roadside trees, on the bark of Quercus cf. robur, 30 May 2022, leg. R. Szymczyk, det. E. A. Ossowska (UGDA L-61375; GenBank OR509533); (3) Miradouro do Rabaçal, 32.756111°N, 17.131111°W, road in Laurel and Juniper forest, saxicolous, 02 June 2022, leg. R. Szymczyk, det E. A. Ossowska (UGDA L-61386; GenBank OR509536).
Parmelia sulcata Taylor
Parmelia sulcata is characterized by sublinear lobes (Figure 6A, Figure 6B), with marginal and laminal soralia (Figure 6B, Figure 6C), simple to squarrose rhizines (Figure 6D), and the presence of salazinic acid in the medulla (see also Hale, 1987; Ossowska, 2021; Thell et al., 2011). Morphologically and chemically similar species are P. barrenoae Divakar, M. C. Molina & A. Crespo, P. encryptata and P. asiatica A. Crespo & Divakar (Divakar et al., 2005; Lumbsch et al., 2011; Molina et al., 2011). From P. encryptata the species differs only in the nucITS rDNA sequence (see notes under that species). The other two taxa differ from P. sulcata in the distribution of the soredia, the lobe shape and width and, the rhizines. Parmelia barrenoae has broad and overlapping lobes, soralia are laminal and like fissures in the upper cortex and rhizines are simple to furcate (Divakar et al., 2005; Hodkinson et al., 2010). In P. asiatica, on the other hand, soralia are predominantly circular and semicircular, terminal, or marginal, while the lobes are narrow and sublinear (Lishtva et al., 2013; Lumbsch et al., 2011). Both taxa have rarely been observed, although some recent data suggest that they may be more common (Ossowska, 2023).
Figure 6
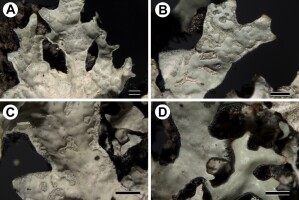
Morphology of Parmelia sulcata from Chile. (A) and (B) sublinear lobes with marginal and laminal soralia (6190 & 6643); (C) marginal and laminal soredia (6643); (D) lobes with abundant, squarrose rhizines (6190). Scale bars: 1 mm.
A diagnostic feature that has been suggested by various authors as crucial in the identification of sorediate Parmelia species is the shape of rhizines (Divakar et al., 2005; Molina et al., 2011; Ossowska & Kukwa, 2016), as P. barrenoae has simple to furcate rhizines while P. sulcata, P. asiatica, and P. encryptata simple to squarrose. However, according to Hodkinson et al. (2010) and Ossowska (2021), the rhizine shape may be confusing, which agrees with our observations on specimens from Chile. In one specimen (leg. Schiefelbein 6615), we observed a dominance of simple rhizines, with only a few squarrose, distributed in the central part of the thallus. At first sight, we assumed that this specimen represented P. encryptata or P. barrenoae. However, the molecular data confirmed that the sample represents P. sulcata. Therefore, caution is needed in the examination of rhizines in sorediate Parmelia specimens, and they need to be checked in several thallus parts, not only in marginal areas.
We obtained nucITS rDNA sequences from three specimens collected in Chile, two of which were identical to P. sulcata sequences (MN387108 and MN654572) deposited by Singh et al. (2019) and Mark et al. (2020). One specimen (leg. Schiefelbein 6615) showed only 97% similarity to the P. sulcata sequence (MN654566). Unfortunately, this sequence was of poor quality, with too many unspecific positions, so we did not include it in further analyses.
To better understand the sequence variation from Chile with other P. sulcata samples, we constructed a haplotype network (Figure 7). To do this, we used most of the nucITS rDNA sequences of P. sulcata deposited in GenBank (Table 1). In total, we found seventeen P. sulcata nucITS rDNA haplotypes (Figure 7), with differences usually in one or two nucleotide positions. The exception is sequence KT625520 from Canada (as haplotype no. 8 in Figure 7). On the tree presented by Molina et al. (2017), this sequence forms a separate clade, sister to P. sulcata. It may represent a new species, but this hypothesis requires further study.
Figure 7
Haplotype network showing relationships among ITS haplotypes of Parmelia sulcata. The new sequences from Chile are marked as red dots.
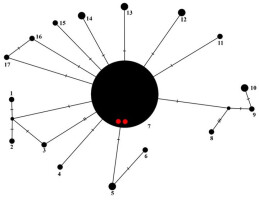
All Parmelia sulcata sequences from Chile represent the most common haplotype (no. 7, Figure 7), also found in specimens from Europe, Asia, and North America (Table 1).
Parmelia sulcata belongs to the widespread Parmelia species and is common, especially in Europe (see Ossowska, 2021 and literature cited therein), although not all records are molecularly confirmed. In Chile this taxon was reported by Hale (1987) from the Magallanes and Stenroos (1991) from Tierra del Fuego, however, the molecular evidence for the occurrence of this species there was only provided by Thell et al. (2002) and Feuerer and Thell (2002) from the department Magallanes y Antártica Chilena. Here, we present a further three records of P. sulcata from Chile supported by molecular data (Figure 8).
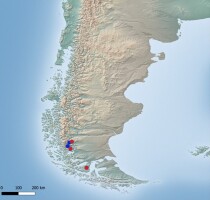
Figure 8
Distribution map of Parmelia sulcata in South America (only records confirmed by molecular data). Blue dots – localities according to literature data, red dots – new localities (base map provided by Natural Earth, www.naturalearthdata.com).
Specimens examined: Chile, (1) Dept. Magallanes y Antártica Chilena, Prov. De Última Esperanza, Torres del Paine, E of national park Torres del Paine, Camino Laguna Azul, 50.856111°S, 72.71056°W, saxicolous, 18 Jan. 2023, leg. U. Schiefelbein 6590, det. E. A. Ossowska (UGDA; GenBank OR509537); (2) Seno Ultima Esperanza, Estancia Perales, 51.542778°S, 72.84361°W, 19 Jan. 2023, leg. U. Schiefelbein 6615, det. E. A. Ossowska (UGDA); (3) Prov. De Magallanes, Punta Arenas, Parrillar Laguna National reserve, forest NW of the camping place along the hiking trail, 53.40444°S, 71.26944°W, 21 Jan. 2023, leg. U. Schiefelbein 6643, det. E. A. Ossowska (UGDA; GenBank OR509538).
. Conclusion
The genus Parmelia includes lichens with large, easily visible thalli, however, their identification can be confusing due to the morphological and chemical similarity between species as well as intraspecific variability. In addition, several new Parmelia species have been described in recent years that can be considered as cryptic or near-cryptic and whose differentiation on the basis of phenetic characteristics may not be correct. We therefore recommend that the identification of Parmelia should be based on the concept of integrative taxonomy. As we show here, quick tools such as a haplotype network or a BLAST search are sufficient for simple taxonomic identification.

