OSMAC Strategy Integrated with Molecular Networking for Accessing Griseofulvin Derivatives from Endophytic Fungi of Moquiniastrum polymorphum (Asteraceae)
Abstract
:1. Introduction
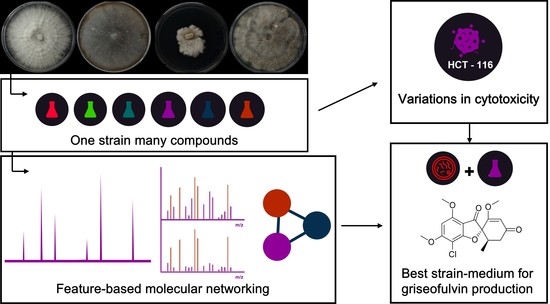
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Isolation, Culture and Extraction, Selection Criteria and Identification of Endophytic Fungi Strains
3.3. Fermentation and Extraction of Strains to “One Strain Many Compounds” (OSMAC) Approach and Feature-Based Molecular Networking (FBMN) Data Processing
3.4. Isolation, Identification, and Quantification of Griseofulvin and 7-Dechlorogriseofulvin
3.5. Cytotoxicity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Sancho, G.; Funk, V.A.; Roque, N. Moquiniastrum (Gochnatieae, Asteraceae): Disentangling the paraphyletic Gochnatia. Phytotaxa 2013, 147, 26–34. [Google Scholar] [CrossRef] [Green Version]
- Martins, G.G.; Lívero, F.A.R.; Stolf, A.M.; Kopruszinski, C.M.; Cardoso, C.C.; Beltrame, O.C.; Queiroz-Telles, J.E.; Strapasson, R.L.B.; Stefanello, M.E.A.; Oude-Elferink, R.; et al. Sesquiterpene lactones of Moquiniastrum polymorphum subsp. floccosum have antineoplastic effects in Walker-256 tumor-bearing rats. Chem. Biol. Interact. 2015, 228, 46–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bueno, N.R.; Castilho, R.O.; Brito, R.; Pott, A.; Pott, V.J.; Scheidt, G.N.; Batista, S. Medicinal plants used by the Kaiowá and Guarani indigenous populations in the Caarapó Reserve, Mato Grosso do Sul, Brazil. Acta Bot. Bras. 2005, 19, 39–44. [Google Scholar] [CrossRef] [Green Version]
- Bohlmann, F.; Zdero, C.; Schmeda-Hirschmann, G.; Jakupovic, J.; Dominguez, X.A.; King, R.M.; Robinson, H. Dimeric guaianolides and other constituents Gochnatia species. Phytochemistry 1986, 25, 1175–1178. [Google Scholar] [CrossRef]
- Catalan, C.A.N.; Vega, M.I.; Lopez, M.E. Coumarins and a kaurane from Gochnatia polymorpha ssp. polymorpha from paraguay. Biochem. Syst. Ecol. 2003, 31, 417–422. [Google Scholar] [CrossRef]
- Sacilotto, A.C.B.C.; Vichnewski, W.; Herz, W. Ent-kaurene diterpenes from Gochnatia polymorpha var. polymorpha. Phytochemistry 1997, 44, 659–661. [Google Scholar] [CrossRef]
- Silva, L.B.; Strapasson, R.L.B.; Riva, D.; Salvador, M.J.; Stefanello, M.É.A. Triterpenes from the flowers of Gochnatia polymorpha subsp. floccosa. Rev. Bras. Farmacogn. 2011, 21, 556–559. [Google Scholar] [CrossRef] [Green Version]
- Piornedo, R.; Souza, P.; Élida, M.; Stefanello, A.; Lauriano, R.; Strapasson, B.; Roberto, A.; Aparecida, C.; Kassuya, L. Anti-inflammatory activity of extracts and 11,13-dihydrozaluzanin C from Gochnatia polymorpha ssp. floccosa trunk bark in mice. J. Ethnopharmacol. 2011, 133, 1077–1084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, M.; Chen, L.; Xin, H.L.; Zheng, C.J.; Rahman, K.; Han, T.; Qin, L.P. A friendly relationship between endophytic fungi and medicinal plants: A systematic review. Front. Microbiol. 2016, 7, 906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.W.; Song, Y.C.; Tan, R.X. Biology and chemistry of endophytes. Nat. Prod. Rep. 2006, 23, 753–771. [Google Scholar] [CrossRef]
- Redman, R.S.; Freeman, S.; Clifton, D.R.; Morrel, J.; Brown, G.; Rodriguez, R.J. Biochemical analysis of plant protection afforded by a nonpathogenic endophytic mutant of Colletotrichum magna. Plant Physiol. 1999, 119, 795–804. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, R.; Redman, R. More than 400 million years of evolution and some plants still can’t make it on their own: Plant stress tolerance via fungal symbiosis. J. Exp. Bot. 2008, 59, 1109–1114. [Google Scholar] [CrossRef]
- Aly, A.H.; Debbab, A.; Proksch, P. Fungal endophytes: Unique plant inhabitants with great promises. Appl. Microbiol. Biotechnol. 2011, 90, 1829–1845. [Google Scholar] [CrossRef] [PubMed]
- Kusari, S.; Hertweck, C.; Spiteller, M. Chemical ecology of endophytic fungi: Origins of secondary metabolites. Chem. Biol. 2012, 19, 792–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Momesso, L.S.; Kawano, C.Y.; Ribeiro, P.H.; Nomizo, A.; Goldman, G.H.; Pupo, M.T. Chaetoglobosinas produzidas por Chaetomium globosum, fungo endofítico associado a Viguiera robusta Gardn. (Asteraceae). Quím. Nova 2008, 31, 1680–1685. [Google Scholar] [CrossRef] [Green Version]
- Gallo, M.B.C.; Chagas, F.O.; Almeida, M.O.; Macedo, C.C.; Cavalcanti, B.C.; Barros, F.W.A.; De Moraes, M.O.; Costa-Lotufo, L.V.; Pessoa, C.; Bastos, J.K.; et al. Endophytic fungi found in association with Smallanthus sonchifolius (Asteraceae) as resourceful producers of cytotoxic bioactive natural products. J. Basic Microbiol. 2009, 49, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Elkhayat, E.S.; Ibrahim, S.R.M.; Mohamed, G.A.; Ross, S.A. Terrenolide S, a new antileishmanial butenolide from the endophytic fungus Aspergillus terreus. Nat. Prod. Res. 2016, 30, 814–820. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Boruta, T. Uncovering the repertoire of fungal secondary metabolites: From Fleming’s laboratory to the international space station. Bioengineered 2018, 9, 12–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Ariantari, N.P.; Daletos, G.; Mándi, A.; Kurtán, T.; Müller, W.E.G.; Lin, W.; Ancheeva, E.; Proksch, P. Expanding the chemical diversity of an endophytic fungus Bulgaria inquinans, an ascomycete associated with mistletoe, through an OSMAC approach. RSC Adv. 2019, 9, 25119–25132. [Google Scholar] [CrossRef] [Green Version]
- Koul, M.; Singh, S. Penicillium spp. prolific producer for harnessing cytotoxic secondary metabolites. Anti-Cancer Drugs 2017, 28, 11–30. [Google Scholar] [CrossRef] [PubMed]
- Petersen, A.B.; Rønnest, M.H.; Larsen, T.O.; Clausen, M.H. The chemistry of griseofulvin. Chem. Rev. 2014, 114, 12088–12107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odds, F.C. Antifungal agents: Their diversity and increasing sophistication. Mycologist 2003, 17, 51–55. [Google Scholar] [CrossRef]
- Chan, Y.C.; Friedlander, S.F. New treatments for tinea capitis. Curr. Opin. Infect. Dis. 2004, 17, 97–103. [Google Scholar] [CrossRef]
- Grisham, L.M.; Wilson, L.; Bensch, K.G. Antimitotic action of griseofulvin does not involve disruption of microtubules. Nature 1973, 244, 294–296. [Google Scholar] [CrossRef] [PubMed]
- Panda, D.; Rathinasamy, K.; Santra, M.K.; Wilson, L. Kinetic suppression of microtubule dynamic instability by giseofulvin implications for its possible use in the treatment of cancer. Proc. Natl. Acad. Sci. USA 2005, 102, 9878–9883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uen, Y.H.; Liu, D.Z.; Weng, M.S.; Ho, Y.S.; Lin, S.Y. NF-kappaB pathway is involved in griseofulvin-induced G2/M arrest and apoptosis in HL-60 cells. J. Cell. Biochem. 2007, 101, 1165–1175. [Google Scholar] [CrossRef]
- Ho, Y.S.; Dun, J.S.; Jeng, J.H.; Wang, Y.J.; Liang, Y.C.; Lin, C.H.; Tseng, C.J.; Yu, C.F.; Chen, R.J.; Lin, J.K. Griseofulvin potentiates antitumorigenesis effects of nocodazole through induction of apoptosis and G2/M cell cycle arrest in human colorectal cancer cells. Int. J. Cancer 2001, 91, 393–401. [Google Scholar] [CrossRef]
- Zhong, N.; Chen, H.; Zhao, Q.; Wang, H.; Yu, X.; Eaves, A.M. Effects of griseofulvin on apoptosis through caspase-3- and caspase-9-dependent pathways in K562 leukemia cells: An in vitro study. Curr. Ther. Res. Clin. Exp. 2010, 71, 384–397. [Google Scholar] [CrossRef] [Green Version]
- Rebacz, B.; Larsen, T.O.; Clausen, M.H.; Ronnest, M.H.; Loffler, H.; Ho, A.D.; Krämer, A. Identification of griseofulvin as an inhibitor of centrosomal clustering in a phenotype-based screen. Cancer Res. 2007, 67, 6342–6350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J. Fungal DNA barcoding: Review. Genome 2016, 932, 913–932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Mello, J.P.F.; Macdonald, A.M.C. Mycotoxins. Anim. Feed Sci. Technol. 1997, 69, 155–166. [Google Scholar] [CrossRef]
- Chapla, V.M.; Zeraik, M.L.; Ximenes, V.F.; Zanardi, L.M.; Lopes, M.N.; Cavalheiro, A.J.; Silva, D.H.S.; Young, M.C.M.; Da Fonseca, L.M.; Bolzani, V.S.; et al. Bioactive secondary metabolites from Phomopsis sp., an endophytic fungus from Senna spectabilis. Molecules 2014, 19, 6597–6608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Z.; Cai, X.; Shao, C.; She, Z.; Xia, X.; Chen, Y.; Yang, J.; Zhou, S.; Lin, Y. Chemistry and weak antimicrobial activities of phomopsins produced by mangrove endophytic fungus Phomopsis sp. ZSU-H76. Phytochemistry 2008, 69, 1604–1608. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.; Yan, X.; Lin, X.; Huang, Y.; Zheng, Z.; Song, S.; Lu, C.; Shen, Y. Chemical constituents of the endophytic fungal strain Phomopsis sp. NXZ-05 of Camptotheca acuminata. Helv. Chim. Acta 2007, 90, 1811–1817. [Google Scholar] [CrossRef]
- Santos, A.L.; Ionta, M.; Horvath, R.; Soares, M.G.; Medeiros, L.S.; Uemi, M.; Kawafune, E.S.; Tangerina, M.M.P.; Ferreira, M.J.P.; Sartorelli, P. Molecular network for accessing polyketide derivatives from Phomopsis sp., an endophytic fungus of Casearia arborea (Salicaceae). Phytochem. Lett. 2021, 42, 1–7. [Google Scholar] [CrossRef]
- Bills, G.F.; González-Menéndez, V.; Martín, J.; Platas, G.; Fournier, J.; Peršoh, D.; Stadler, M. Hypoxylon pulicicidum sp. nov. (Ascomycota, Xylariales), a pantropical insecticide-producing endophyte. PLoS ONE 2012, 7, e46687. [Google Scholar] [CrossRef]
- Liang, H.-Q.; Zhang, D.-W.; Guo, S.-X.; Yu, J. Two new tetracyclic triterpenoids from the endophytic fungus Hypoxylon sp. 6269. J. Asian Nat. Prod. Res. 2018, 20, 951–956. [Google Scholar] [CrossRef] [PubMed]
- Porritt, A.E. The Discovery and Development of Penicillin; Royal Society of Chemistry: London, UK, 1999. [Google Scholar]
- Nothias, L.F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Brian, P.W.; Curtis, P.J.; Hemming, H.G. A substance causing abnormal development of fungal hyphae produced by Penicillium janczewskii Zal.: I. Biological assay, production and isolation of ‘curling factor’. Trans. Br. Mycol. Soc. 1946, 29, 173–187. [Google Scholar] [CrossRef]
- Grove, J.F.; McGowan, J.C. Identity of griseofulvin and ‘curling-factor’. Nature 1947, 160, 574. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, M.; Sanchis, V.; Mateo, R.; Hernández, E. Detection and quantification of patulin and griseofulvin by high pressure liquid chromatography in different strains of Penicillium griseofulvum Dierckx. Mycotoxin Res. 1988, 4, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Dorey, M.J.; Mitchell, I.L.S.; Rule, D.W.; Walker, C. Production of Griseofulvin. U.S. Patent 3,069,329, 12 June 1962. [Google Scholar]
- Calvo, A.M.; Wilson, R.A.; Bok, J.W.; Keller, N.P. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 2002, 66, 447–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, B.; Parrot, D.; Blümel, M.; Labes, A.; Tasdemir, D. Influence of OSMAC-based cultivation in metabolome and anticancer activity of fungi associated with the brown alga Fucus vesiculosus. Mar. Drugs 2019, 17, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tudzynski, B. Nitrogen regulation of fungal secondary metabolism in fungi. Front. Microbiol. 2014, 5, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brakhage, A.A. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 2013, 11, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Hemphill, C.F.P.; Sureechatchaiyan, P.; Kassack, M.U.; Orfali, R.S.; Lin, W.; Daletos, G.; Proksch, P. OSMAC approach leads to new fusarielin metabolites from Fusarium tricinctum. J. Antibiot. 2017, 70, 726–732. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Cafêu, M.C.; Silva, G.H.; Teles, H.L.; Bolzani, V.S.; Araújo, A.R.; Young, M.C.M.; Pfenning, L.H. Substâncias antifúngicas de Xylaria sp., um fungo endofítico isolado de Palicourea marcgravii (Rubiaceae). Quim. Nova 2005, 28, 991–995. [Google Scholar] [CrossRef] [Green Version]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Tamayose, C.I.; Santos, E.A.; Roque, N.; Costa-Lotufo, L.V.; Ferreira, M.J.P. Caffeoylquinic acids: Separation method, antiradical properties and cytotoxicity. Chem. Biodivers. 2019, 16, e1900093. [Google Scholar] [CrossRef] [PubMed]
- Tangerina, M.M.P.; Furtado, L.C.; Leite, V.M.B.; Bauermeister, A.; Velasco-Alzate, K.; Jimenez, P.C.; Garrido, L.M.; Padilla, G.; Lopes, N.P.; Costa-Lotufo, L.V.; et al. Metabolomic study of marine Streptomyces sp.: Secondary metabolites and the production of potential anticancer compounds. PLoS ONE 2020, 15, e0244385. [Google Scholar] [CrossRef] [PubMed]


| % of Griseofulvin 2 | % of 7-Dechlorogriseofulvin 2 | |||||
|---|---|---|---|---|---|---|
| Culture Media 1 | 611 | 620 | 658 | 611 | 620 | 658 |
| CZK | 3.80 | 0.80 | 1.30 | 1.02 | 0.29 | 0.55 |
| MEP | 7.24 | 7.61 | 3.63 | 2.94 | 1.78 | 0.63 |
| PDB | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| PDA | 0.0 | 7.03 | 0.52 | 0.0 | 0.37 | 0.09 |
| R | 1.34 | 2.25 | 0.26 | 0.58 | 0.28 | 0.45 |
| WKM | 0.37 | 7.74 | 3.13 | 1.61 | 4.54 | 0.61 |
| Crude Extracts Obtained from Culture Media 2 | ||||||
|---|---|---|---|---|---|---|
| Strains 3 | CZK | MEP | PDB | PDA | R | WKM |
| 611 | 43.5 ± 8.3 | 65.8 ± 2.6 | 58.7 ± 4.1 | 8.6 ± 11.0 | 62.8 ± 3.0 | 62.3 ± 1.4 |
| 620 | 36.3 ± 5.9 | 68.7 ± 0.7 | 61.1 ± 3.5 | 44.8 ± 7.2 | 71.4 ± 5.0 | 30.6 ± 2.5 |
| 658 | 48.2 ± 7.9 | 63.4 ± 4.3 | 59.6 ± 5.5 | 62.0 ± 5.4 | 64.4 ± 4.9 | 53.7 ± 6.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farinella, V.F.; Kawafune, E.S.; Tangerina, M.M.P.; Domingos, H.V.; Costa-Lotufo, L.V.; Ferreira, M.J.P. OSMAC Strategy Integrated with Molecular Networking for Accessing Griseofulvin Derivatives from Endophytic Fungi of Moquiniastrum polymorphum (Asteraceae). Molecules 2021, 26, 7316. https://doi.org/10.3390/molecules26237316
Farinella VF, Kawafune ES, Tangerina MMP, Domingos HV, Costa-Lotufo LV, Ferreira MJP. OSMAC Strategy Integrated with Molecular Networking for Accessing Griseofulvin Derivatives from Endophytic Fungi of Moquiniastrum polymorphum (Asteraceae). Molecules. 2021; 26(23):7316. https://doi.org/10.3390/molecules26237316
Chicago/Turabian StyleFarinella, Victor F., Eunizinis S. Kawafune, Marcelo M. P. Tangerina, Helori V. Domingos, Leticia V. Costa-Lotufo, and Marcelo J. P. Ferreira. 2021. "OSMAC Strategy Integrated with Molecular Networking for Accessing Griseofulvin Derivatives from Endophytic Fungi of Moquiniastrum polymorphum (Asteraceae)" Molecules 26, no. 23: 7316. https://doi.org/10.3390/molecules26237316