Lichen-Derived Diffractaic Acid Inhibited Dengue Virus Replication in a Cell-Based System
Abstract
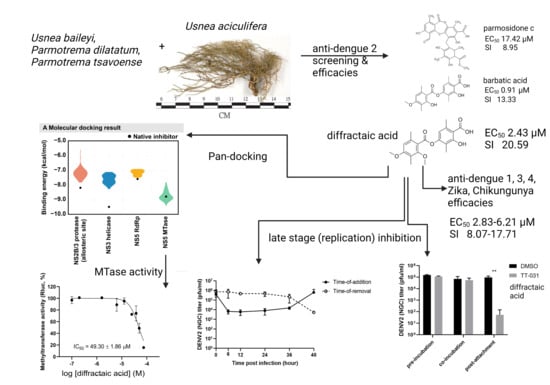
:1. Introduction
2. Results
2.1. Preliminary Screening, Efficacies, and Cytotoxicities in DENV-2-Infected Cells
2.2. Cytotoxicities and Efficacies of Diffractaic Acid in other Human-Derived Cell Lines and Mosquito-Borne Viruses
2.3. Diffractaic Acid Inhibits DENV2 after Entry and Interferes with Viral Replication
3. Discussion
4. Materials and Methods
4.1. Extraction, Purification, and Identification of Lichen Depsidones and Depsides
4.2. Cells and Viruses
4.3. Efficacy Test
4.4. Time Course Assay
4.5. Cytotoxicity Test
4.6. Attachment Inhibition Study
4.7. Fusion Inhibition Assay
4.8. RNA Extraction and Quantitative RT-PCR (RT-qPCR)
4.9. Western Blotting
4.10. Molecular Docking Study
4.11. Dengue Methyltransferase (MTase) Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Vicente, C.R.; Herbinger, K.-H.; Fröschl, G.; Malta Romano, C.; de Souza Areias Cabidelle, A.; Cerutti Junior, C. Serotype influences on dengue severity: A cross-sectional study on 485 confirmed dengue cases in Vitória, Brazil. BMC Infect. Dis. 2016, 16, 320. [Google Scholar] [CrossRef] [Green Version]
- Fried, J.R.; Gibbons, R.V.; Kalayanarooj, S.; Thomas, S.J.; Srikiatkhachorn, A.; Yoon, I.K.; Jarman, R.G.; Green, S.; Rothman, A.L.; Cummings, D.A. Serotype-specific differences in the risk of dengue hemorrhagic fever: An analysis of data collected in Bangkok, Thailand from 1994 to 2006. PLoS Negl. Trop. Dis. 2010, 4, e617. [Google Scholar] [CrossRef] [Green Version]
- Ooi, E.E. Repurposing Ivermectin as an Anti-dengue Drug. Clin. Infect. Dis. 2020, 72, e594–e595. [Google Scholar] [CrossRef]
- Crawford, S.D. Lichens Used in Traditional Medicine. In Lichen Secondary Metabolites: Bioactive Properties and Pharmaceutical Potential; Ranković, B., Ed.; Springer International Publishing: Cham, Germany, 2015; pp. 27–80. [Google Scholar]
- Gautam, A.K.; Yadav, D.; Singh, P.K.; Bhagyawant, S.S.; Jin, J.-O. Lichen: A comprehensive review on Lichens as a natural sources exploring nutritional and biopharmaceutical benefits. Prog. Nutr. 2021, 23, e2021153. [Google Scholar] [CrossRef]
- Adenubi, O.T.; Famuyide, I.M.; McGaw, L.J.; Eloff, J.N. Lichens: An update on their ethnopharmacological uses and potential as sources of drug leads. J. Ethnopharmacol. 2022, 298, 115657. [Google Scholar] [CrossRef]
- Ranković, B.; Kosanić, M. Lichens as a potential source of bioactive secondary metabolites. In Lichen Secondary Metabolites; Springer: Cham, Switzerland, 2019; pp. 1–29. [Google Scholar]
- Lohézic-Le Dévéhat, F.; Tomasi, S.; Elix, J.A.; Bernard, A.; Rouaud, I.; Uriac, P.; Boustie, J. Stictic acid derivatives from the lichen Usnea articulata and their antioxidant activities. J. Nat. Prod. 2007, 70, 1218–1220. [Google Scholar] [CrossRef]
- Melo, M.G.D.; dos Santos, J.P.A.; Serafini, M.R.; Caregnato, F.F.; de Bittencourt Pasquali, M.A.; Rabelo, T.K.; da Rocha, R.F.; Quintans, L., Jr.; de Souza Araújo, A.A.; da Silva, F.A. Redox properties and cytoprotective actions of atranorin, a lichen secondary metabolite. Toxicol. Vitr. 2011, 25, 462–468. [Google Scholar] [CrossRef] [Green Version]
- Ranković, B.; Mišić, M. The antimicrobial activity of the lichen substances of the lichens Cladonia furcata, Ochrolechia androgyna, Parmelia caperata and Parmelia conspresa. Biotechnol. Biotechnol. Equip. 2008, 22, 1013–1016. [Google Scholar] [CrossRef] [Green Version]
- Varughese, T.; Rios, N.; Higginbotham, S.; Arnold, A.E.; Coley, P.D.; Kursar, T.A.; Gerwick, W.H.; Rios, L.C. Antifungal depsidone metabolites from Cordyceps dipterigena, an endophytic fungus antagonistic to the phytopathogen Gibberella fujikuroi. Tetrahedron Lett. 2012, 53, 1624–1626. [Google Scholar] [CrossRef] [Green Version]
- Silva, H.; Aires, A.; Soares, C.; Sá, J.; Martins, M.; Albuquerque, M.; Silva, T.; Brayner, F.; Alves, L.; Melo, A. Barbatic acid from Cladia aggregata (lichen): Cytotoxicity and in vitro schistosomicidal evaluation and ultrastructural analysis against adult worms of Schistosoma mansoni. Toxicol. Vitr. 2020, 65, 104771. [Google Scholar] [CrossRef]
- Reddy, S.D.; Siva, B.; Kumar, K.; Babu, V.; Sravanthi, V.; Boustie, J.; Nayak, V.L.; Tiwari, A.K.; Rao, C.; Sridhar, B. Comprehensive Analysis of Secondary Metabolites in Usnea longissima (Lichenized Ascomycetes, Parmeliaceae) Using UPLC-ESI-QTOF-MS/MS and Pro-Apoptotic Activity of Barbatic Acid. Molecules 2019, 24, 2270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harikrishnan, A.; Veena, V.; Lakshmi, B.; Shanmugavalli, R.; Theres, S.; Prashantha, C.; Shah, T.; Oshin, K.; Togam, R.; Nandi, S. Atranorin, an antimicrobial metabolite from lichen Parmotrema rampoddense exhibited in vitro anti-breast cancer activity through interaction with Akt activity. J. Biomol. Struct. Dyn. 2020, 39, 1248–1258. [Google Scholar] [CrossRef] [PubMed]
- Dongmo Zeukang, R.; Siwe-Noundou, X.; Tagatsing Fotsing, M.; Tabopda Kuiate, T.; Mbafor, J.T.; Krause, R.W.; Choudhary, M.I.; Atchadé, A.d.T. Cordidepsine is A Potential New Anti-HIV Depsidone from Cordia millenii, Baker. Molecules 2019, 24, 3202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odimegwu, D.C. Low-dose Sekikaic acid modulates host immunity and protects cells from Respiratory Syncytial Virus infection. Biotechnol. J. Int. 2018, 21, 1–10. [Google Scholar] [CrossRef]
- Vu, T.H.; Le Lamer, A.-C.; Lalli, C.; Boustie, J.; Samson, M.; Lohézic-Le Dévéhat, F.; Le Seyec, J. Depsides: Lichen metabolites active against hepatitis C virus. PLoS ONE 2015, 10, e0120405. [Google Scholar]
- Yao, Y.; Huo, T.; Lin, Y.-L.; Nie, S.; Wu, F.; Hua, Y.; Wu, J.; Kneubehl, A.R.; Vogt, M.B.; Rico-Hesse, R.; et al. Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease. J. Am. Chem. Soc. 2019, 141, 6832–6836. [Google Scholar] [CrossRef]
- Byrd, C.M.; Grosenbach, D.W.; Berhanu, A.; Dai, D.; Jones, K.F.; Cardwell, K.B.; Schneider, C.; Yang, G.; Tyavanagimatt, S.; Harver, C.; et al. Novel benzoxazole inhibitor of dengue virus replication that targets the NS3 helicase. Antimicrob. Agents Chemother. 2013, 57, 1902–1912. [Google Scholar] [CrossRef] [Green Version]
- Noble, C.G.; Lim, S.P.; Chen, Y.-L.; Liew, C.W.; Yap, L.; Lescar, J.; Shi, P.-Y. Conformational flexibility of the Dengue virus RNA-dependent RNA polymerase revealed by a complex with an inhibitor. J. Virol. 2013, 87, 5291–5295. [Google Scholar] [CrossRef] [Green Version]
- Lafreniere, M.A.; Desrochers, G.F.; Mekbib, K.; Pezacki, J.P. An affinity-based probe for methyltransferase enzymes based on sinefungin. Can. J. Chem. 2017, 95, 1059–1063. [Google Scholar] [CrossRef]
- Boonyasuppayakorn, S.; Saelee, T.; Visitchanakun, P.; Leelahavanichkul, A.; Hengphasatporn, K.; Shigeta, Y.; Huynh, T.N.T.; Chu, J.J.H.; Rungrotmongkol, T.; Chavasiri, W. Dibromopinocembrin and Dibromopinostrobin Are Potential Anti-Dengue Leads with Mild Animal Toxicity. Molecules 2020, 25, 4154. [Google Scholar] [CrossRef]
- Duong, T.-H.; Chavasiri, W.; Boustie, J. New meta-depsidones and diphenyl ethers from the lichen Parmotrema tsavoense (Krog & Swinscow) Krog & Swinscow, Parmeliaceae. Tetrahedron 2015, 71, 9684–9691. [Google Scholar]
- Emsen, B.; Aslan, A.; Turkez, H.; Joughi, A.T.; Kaya, A. The anti-cancer efficacies of diffractaic, lobaric, and usnic acid: In Vitro inhibition of glioma. J. Cancer Res. Ther. 2018, 14, 941–951. [Google Scholar] [CrossRef]
- Karagoz, I.; Ozaslan, M.; Guler, I.; Uyar, C.; Yalim, T.; Kazanci, U.; Aslan, A.; Cakir, A. In Vivo antitumoral effect of diffractaic acid from lichen metabolites on Swiss albino mice with Ehrlich ascites carcinoma: An experimental study. Int. J. Pharmacol. 2014, 10, 307–314. [Google Scholar] [CrossRef] [Green Version]
- Odabasoglu, F.; Yildirim, O.S.; Aygun, H.; Halici, Z.; Halici, M.; Erdogan, F.; Cadirci, E.; Cakir, A.; Okumus, Z.; Aksakal, B. Diffractaic acid, a novel proapoptotic agent, induces with olive oil both apoptosis and antioxidative systems in Ti-implanted rabbits. Eur. J. Pharmacol. 2012, 674, 171–178. [Google Scholar] [CrossRef]
- Berdy, J.; Boca Raton, F.L. CRC Handbook of Antibiotic Compounds; CRC Press: Boca Raton, FL, USA, 1981; Volume 1. [Google Scholar]
- Baggen, J.; Thibaut, H.J.; Strating, J.R.; van Kuppeveld, F.J. The life cycle of non-polio enteroviruses and how to target it. Nat. Rev. Microbiol. 2018, 16, 368–381. [Google Scholar] [CrossRef]
- Truong, T.L.; Nga, V.T.; Huy, D.T.; Chi, H.B.; Phung, N.K. A new depside from Usnea aciculifera growing in Vietnam. Nat. Prod. Commun. 2014, 9, 1179–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Nguyen, K.; Duong, T.-H.; Nguyen, K.P.P.; Sangvichien, E.; Wonganan, P.; Chavasiri, W. Chemical constituents of the lichen Usnea baileyi (Stirt.) Zahlbr. Tetrahedron Lett. 2018, 59, 1348–1351. [Google Scholar] [CrossRef]
- Devi, A.P.; Duong, T.-H.; Ferron, S.; Beniddir, M.A.; Dinh, M.-H.; Nguyen, V.-K.; Mac, D.-H.; Boustie, J.; Chavasiri, W.; Le Pogam, P. Salazinic Acid-Derived Depsidones and Diphenylethers with α-Glucosidase Inhibitory Activity from the Lichen Parmotrema dilatatum. Planta Med. 2020, 86, 1216–1224. [Google Scholar] [CrossRef]
- Suroengrit, A.; Yuttithamnon, W.; Srivarangkul, P.; Pankaew, S.; Kingkaew, K.; Chavasiri, W.; Boonyasuppayakorn, S. Halogenated chrysins inhibit dengue and zika virus infectivity. Sci. Rep. 2017, 7, 13696. [Google Scholar] [CrossRef] [Green Version]
- Boonyasuppayakorn, S.; Suroengrit, A.; Srivarangkul, P.; Yuttithamnon, W.; Pankaew, S.; Saelee, T.; Prompetchara, E.; Salakij, S.; Bhattarakosol, P. Simplified dengue virus microwell plaque assay using an automated quantification program. J. Virol. Methods 2016, 237, 25–31. [Google Scholar] [CrossRef]
- Kanyaboon, P.; Saelee, T.; Suroengrit, A.; Hengphasatporn, K.; Rungrotmongkol, T.; Chavasiri, W.; Boonyasuppayakorn, S. Cardol triene inhibits dengue infectivity by targeting kl loops and preventing envelope fusion. Sci. Rep. 2018, 8, 16643. [Google Scholar] [CrossRef] [Green Version]
- Kajita, M.; Katayama, H.; Murata, T.; Kai, C.; Hori, M.; Ozaki, H. Canine Distemper Virus Induces Apoptosis Through Caspase-3 and-8 Activation in Vero Cells. J. Vet. Med. Ser. B 2006, 53, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Erbel, P.; Schiering, N.; D’Arcy, A.; Renatus, M.; Kroemer, M.; Lim, S.P.; Yin, Z.; Keller, T.H.; Vasudevan, S.G.; Hommel, U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat. Struct. Mol. Biol. 2006, 13, 372–373. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Sampath, A.; Chao, A.; Wen, D.; Nanao, M.; Chene, P.; Vasudevan, S.G.; Lescar, J. Structure of the Dengue virus helicase/nucleoside triphosphatase catalytic domain at a resolution of 2.4 A. J. Virol. 2005, 79, 10278–10288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benmansour, F.; Trist, I.; Coutard, B.; Decroly, E.; Querat, G.; Brancale, A.; Barral, K. Discovery of novel dengue virus NS5 methyltransferase non-nucleoside inhibitors by fragment-based drug design. Eur. J. Med. Chem. 2017, 125, 865–880. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16 Rev. C.01. 2016. Available online: https://gaussian.com/citation/ (accessed on 20 August 2022).
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hengphasatporn, K.; Kungwan, N.; Rungrotmongkol, T. Binding pattern and susceptibility of epigallocatechin gallate against envelope protein homodimer of Zika virus: A molecular dynamics study. J. Mol. Liq. 2019, 274, 140–147. [Google Scholar] [CrossRef]
- Hengphasatporn, K.; Wilasluck, P.; Deetanya, P.; Wangkanont, K.; Chavasiri, W.; Visitchanakun, P.; Leelahavanichkul, A.; Paunrat, W.; Boonyasuppayakorn, S.; Rungrotmongkol, T.; et al. Halogenated Baicalein as a Promising Antiviral Agent toward SARS-CoV-2 Main Protease. J. Chem. Inf. Model. 2022, 62, 1498–1509. [Google Scholar] [CrossRef]
- BIOVIA. Dassault Systèmes, Discovery Studio Visualizer; BIOVIA: San Diego, CA, USA, 2021. [Google Scholar]
- Boonyasuppayakorn, S.; Padmanabhan, R. Construction of Plasmid, Bacterial Expression, Purification, and Assay of Dengue Virus Type 2 NS5 Methyltransferase. In Dengue: Methods and Protocols; Padmanabhan, R., Vasudevan, S.G., Eds.; Springer: New York, NY, USA, 2014; pp. 361–373. [Google Scholar]
- Randolph, V.B.; Stollar, V. Low pH-induced cell fusion in flavivirus-infected Aedes albopictus cell cultures. J. Gen. Virol. 1990, 71, 1845–1850. [Google Scholar] [CrossRef]




| Code | IUPAC Name | Common Name | Biological Source | Viral Inhibition | Cell Viability | SI | ||
|---|---|---|---|---|---|---|---|---|
| 1 | Depsides | DENV2 NGC (%) 1 | EC50 (µM) 2 | Vero (%) 1 | CC50 (µM) 2 | |||
| 1a | 4-((2,4-dimethoxy-3,6-dimethylbenzoyl)oxy)-2-hydroxy-3,6-dimethylbenzoic acid | Diffractaic acid | Usnea aciculifera | 99.98 ± 0.04 | DENV2 (NGC) 2.43 ± 0.19 DENV2 (16681) 3.89 ± 0.07 DENV1 (16007) 4.57 ± 0.26 DENV3 (16562) 4.90 ± 0.49 DENV4 (1036) 4.43 ± 0.58 ZIKV (SV0127/14) 2.83 ± 0.50 CHIKV (ECSA) 6.21 ± 0.69 EV-A71 (BRCR) 19.93 ± 1.94 | 114.72 ± 5.88 | Vero 50.13 ± 7.45 Huh-7 >100 HepG2 >100 HEK-293 70.44 ± 0.33 THP-1 49.60 ± 1.06 RD 64.34 ± 5.04 | DENV2 (NGC) 20.59 DENV2 (16681) >25 DENV1 10.97 DENV3 10.23 DENV4 11.33 ZIKV 17.71 CHIKV 8.07 EV-A71 3.32 |
| 1b | 2-hydroxy-4-((2-hydroxy-4-methoxy-3,6-dimethylbenzoyl)oxy)-3,6-dimethylbenzoic acid | Barbatic acid | Usnea aciculifera | 99.99 ± 0.02 | DENV2 (NGC) 0.91 ± 0.15 | 113.31 ± 7.29 | Vero 12.10 ± 0.38 | DENV2 (NGC) 13.33 |
| 2, 3 | Depsidones | |||||||
| 2a | 6-(2,4-dihydroxy-5-(methoxycarbonyl)-3,6-dimethylben-zyl)-4-formyl-3,7-dihydroxy-1,9-dimethyl-11-oxo-11H-dibenzo[b,e][1,4]dioxepine-8-carboxylic acid | Parmosidone C | Parmotrema tsavoense | 96.48 ± 2.90 | DENV2 (NGC) 17.42 ± 3.21 | 123.58 ± 17.10 | Vero 155.83 ± 7.77 | DENV2 (NGC) 8.95 |
| 2b | 4-formyl-3,8-dihydroxy-1,6-dimethyl-11-oxo-11H-dibenzo[b,e][1,4]dioxepine-7-carboxylic acid | Subvirensic acid | Usnea baileyi | Not inhibited | 114.15 ± 15.82 | |||
| 2c | 4-formyl-3,8-dihydroxy-9-(methoxymethyl)-1,6-dimethyl-11-oxo-11H-dibenzo[b,e][1,4]dioxepine-7-carboxylic acid | 9′-O-methylprotocetraric acid | Usnea baileyi | Not inhibited | 102.53 ± 3.32 | |||
| 3a | (11-formyl-1,4,10-trihydroxy-8-methyl-3,7-dioxo-1,3-dihydro-7H-benzo[6,7][1,4]dioxepino[2,3-e]isobenzofuran-5-yl)methyl acetate | Galbinic Acid | Parmotrema dilatatum | 80.50 ± 4.85 | 112.03 ± 3.55 | |||
| 3b | 1,4,10-trihydroxy-5,8-dimethyl-3,7-dioxo-1,3-dihy-dro-7H-benzo[6,7][1,4]dioxepino[2,3-e]isobenzofuran-11-carbaldehyde | Norstictic acid | Usnea aciculifera | 78.00 ± 7.55 | 111.18 ± 5.63 | |||
| 3c | 1,4-dihydroxy-10-methoxy-5,8-dimethyl-3,7-dioxo-1,3-dihydro-7H-benzo[6,7][1,4]dioxepino[2,3-e]isobenzofuran-11-carbaldehyde | Stictic acid | Usnea aciculifera | Not inhibited | 108.02 ± 4.34 | |||
| 3d | 1,4,10-trihydroxy-8-methyl-3,7-dioxo-5-(2,4,6-trihydroxyben-zyl)-1,3-dihydro-7H-benzo[6,7][1,4]dioxepino[2,3-e]isobenzofuran-11-carbaldehyde | Parmosidone F | Parmotrema dilatatum | Not inhibited | 120.97 ± 12.71 | |||
| 3e | 1,4,10-trihydroxy-5-(hydroxymethyl)-8-methyl-3,7-dioxo-1,3-dihydro-7H-benzo[6,7][1,4]dioxepino[2,3-e]isobenzofuran-11-carbaldehyde | Salazinic acid | Parmotrema dilatatum | Not inhibited | 120.10 ± 6.87 | |||
| 3f | 4-hydroxy-5-(hydroxymethyl)-1,10-dimethoxy-8-methyl-3,7-dioxo-1,3-dihydro-7H-benzo[6,7][1,4]dioxepino[2,3-e]isobenzofuran-11-carbaldehyde | 8′-O-methylconstictic acid | Usnea baileyi | Not inhibited | 112.63 ± 17.52 | |||
| 3g | 4,11-dihydroxy-1,10-dimethoxy-5,8-dimethyl-7H-benzo[6,7][1,4]dioxepino[2,3-e]isobenzofuran-3,7(1H)-dione | 8′-O-methylmenegazziaic acid | Usnea baileyi | Not inhibited | 122.40 ± 4.58 | |||
| 3h | 4-hydroxy-1,10-dimethoxy-5,8-dimethyl-3,7-dioxo-1,3-dihydro-7H-benzo[6,7][1,4]dioxepino[2,3-e]isobenzofuran-11-carbaldehyde | Methylstictic acid | Usnea baileyi | Not inhibited | 118.00 ± 4.69 | |||
| 3i | 4-hydroxy-11-(hydroxymethyl)-1,10-dimethoxy-5,8-dimethyl-7H-benzo[6,7][1,4]dioxepino[2,3-e]isobenzofuran-3,7(1H)-dion | 8′-O-methylcryptostictic acid | Usnea baileyi | Not inhibited | 108.59 ± 5.75 | |||
| 3j | 1-ethoxy-4-hydroxy-10-methoxy-5,8-dimethyl-3,7-dioxo-1,3-dihydro-7H-benzo[6,7][1,4]dioxepino[2,3-e]isobenzofuran-11-carbaldehyde | 8′-O-ethylstictic acid | Usnea baileyi | Not inhibited | 97.88 ± 5.73 | |||
| 3k | 1,4,11-trihydroxy-10-methoxy-5,8-dimethyl-7H-benzo[6,7][1,4]dioxepino[2,3-e]isobenzofuran-3,7(1H)-dione | Menegazziaic acid | Usnea baileyi | Not inhibited | 92.00 ± 11.41 | |||
| 4 | methyl 2,4-dihydroxy-6-methylbenzoate | Methyl orselinate | Usnea aciculifera | Not inhibited | 106.62 ± 8.05 | |||
| 5 | 5-hydroxy-7-methoxy-6-methylisobenzofuran-1(3H)-one | 7-hydroxy-5-methoxy-6-methylphthalide | Usnea aciculifera | Not inhibited | 119.68 ± 11.66 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loeanurit, N.; Tuong, T.L.; Nguyen, V.-K.; Vibulakhaophan, V.; Hengphasatporn, K.; Shigeta, Y.; Ho, S.X.; Chu, J.J.H.; Rungrotmongkol, T.; Chavasiri, W.; et al. Lichen-Derived Diffractaic Acid Inhibited Dengue Virus Replication in a Cell-Based System. Molecules 2023, 28, 974. https://doi.org/10.3390/molecules28030974
Loeanurit N, Tuong TL, Nguyen V-K, Vibulakhaophan V, Hengphasatporn K, Shigeta Y, Ho SX, Chu JJH, Rungrotmongkol T, Chavasiri W, et al. Lichen-Derived Diffractaic Acid Inhibited Dengue Virus Replication in a Cell-Based System. Molecules. 2023; 28(3):974. https://doi.org/10.3390/molecules28030974
Chicago/Turabian StyleLoeanurit, Naphat, Truong Lam Tuong, Van-Kieu Nguyen, Vipanee Vibulakhaophan, Kowit Hengphasatporn, Yasuteru Shigeta, Si Xian Ho, Justin Jang Hann Chu, Thanyada Rungrotmongkol, Warinthorn Chavasiri, and et al. 2023. "Lichen-Derived Diffractaic Acid Inhibited Dengue Virus Replication in a Cell-Based System" Molecules 28, no. 3: 974. https://doi.org/10.3390/molecules28030974