The Qc5 Allele Increases Wheat Bread-Making Quality by Regulating SPA and SPR
Abstract
:1. Introduction
2. Results
2.1. Phenotype and Segregation Analyses
2.2. Gene Cloning and Expression Analysis
2.3. Protein Structure Analysis
2.4. Processing Quality Analysis
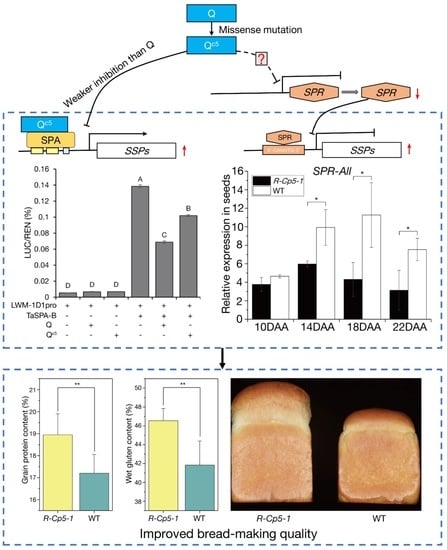
2.5. Mechanisms Underlying the Effect of Qc5 on Processing Quality
2.6. Effect of Qc5 on the Seed Transcriptome
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. DNA Extraction and Segregation Analysis
4.3. RNA Extraction and Gene Expression Analysis
4.4. Gene Cloning
4.5. Structure Prediction
4.6. Processing Quality Analysis
4.7. Reversed-Phase High-Performance Liquid Chromatography (RP-HPLC)
4.8. Subcellular Localization
4.9. Yeast One- and Two-Hybrid Analyses
4.10. Dual-Luciferase Reporter Assay
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nesbitt, M.; Samuel, D. From the staple crop to extinction? The archaeology and history of hulled wheats. In Hulled Wheats; Padulosi, S., Hammer, K., Heller, J., Eds.; International Plant Genetic Resources Institute: Rome, Italy, 1996; Volume 4, pp. 41–100. [Google Scholar]
- Huang, S.; Sirikhachornkit, A.; Su, X.; Faris, J.; Gill, B.; Haselkorn, R.; Gornicki, P. Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. USA 2002, 99, 8133–8138. [Google Scholar] [CrossRef] [Green Version]
- FAOSTAT. FAO Database. Food and Agriculture Organization of the United Nations. Available online: http://www.fao.org/faostat/en/#data (accessed on 26 July 2021).
- Shewry, P.R. Wheat. J. Exp. Bot. 2009, 60, 1537–1553. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.F.; Wei, Y.M.; Yue, Y.W.; Yan, Z.H.; Zheng, Y.L. Biochemical and molecular characterization of gliadins. Mol. Biol. 2006, 40, 713–723. [Google Scholar] [CrossRef]
- Payne, P.I. Genetics of wheat storage proteins and the effect of allelic variation on bread-making quality. Annu. Rev. Physiol. 1987, 38, 141–153. [Google Scholar] [CrossRef]
- Don, C.; Mann, G.; Bekes, F.; Hamer, R.J. HMW-GS affect the properties of glutenin particles in GMP and thus flour quality. J. Cereal Sci. 2006, 44, 127–136. [Google Scholar] [CrossRef]
- Tian, J.; Deng, Z.; Zhang, K.; Yu, H.; Jiang, X.; Li, C. Genetic detection of main quality traits in wheat. In Genetic Analyses of Wheat and Molecular Marker-Assisted Breeding; Science Press: Beijing, China, 2015; Volume 5, pp. 177–350. [Google Scholar]
- Rubio-Somoza, I.; Martinez, M.; Abraham, Z.; Diaz, I.; Carbonero, P. Ternary complex formation between HvMYBS3 and other factors involved in transcriptional control in barley seeds. Plant J. 2006, 47, 269–281. [Google Scholar] [CrossRef]
- Verdier, J.; Thompson, R.D. Transcriptional regulation of storage protein synthesis during dicotyledon seed filling. Plant Cell Physiol. 2008, 49, 1263–1271. [Google Scholar] [CrossRef] [Green Version]
- Xi, D.M.; Zheng, C.C. Transcriptional regulation of seed storage protein genes in Arabidopsis and cereals. Seed Sci. Res. 2011, 21, 247–254. [Google Scholar] [CrossRef]
- Vicente-Carbajosa, J.; Moose, S.P.; Parsons, R.L.; Schmidt, R.J. A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc. Natl. Acad. Sci. USA 1997, 94, 7685–7690. [Google Scholar] [CrossRef] [Green Version]
- Oñate, L.; Vicente-Carbajosa, J.; Lara, P.; Diaz, I.; Carbonero, P. Barley BLZ2, a seed-specific bZIP protein that interacts with BLZ1 in vivo and activates transcription from the GCN4-like motif of B-hordein promoters in barley endosperm. J. Biol. Chem. 1999, 274, 9175–9182. [Google Scholar] [CrossRef] [Green Version]
- Albani, D.; Hammond-Kosack, M.C.; Smith, C.; Conlan, S.; Colot, V.; Holdsworth, M.; Bevan, M.W. The wheat transcriptional activator SPA: A seed-specific bZIP protein that recognizes the GCN4-like motif in the bifactorial endosperm box of prolamin genes. Plant Cell 1997, 9, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Mena, M.; Vicente-Carbajosa, J.; Schmidt, R.J.; Carbonero, P. An endosperm-specific DOF protein from barley, highly conserved in wheat, binds to and activates transcription from the prolamin-box of a native B-hordein promoter in barley endosperm. Plant J. 1998, 16, 53–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz, I.; Martinez, M.; Isabel-LaMoneda, I.; Rubio-Somoza, I.; Carbonero, P. The DOF protein, SAD, interacts with GAMYB in plant nuclei and activates transcription of endosperm-specific genes during barley seed development. Plant J. 2005, 42, 652–662. [Google Scholar] [CrossRef]
- Bäumlein, H.; Nagy, I.; Villarroel, R.; Inzé, D.; Wobus, U. Cis-analysis of a seed protein gene promoter: The conservative RY repeat CATGCATG within the legumin box is essential for tissue-specific expression of a legumin gene. Plant J. 1992, 2, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Risueno, M.A.; González, N.; Díaz, I.; Parcy, F.; Carbonero, P.; Vicente-Carbajosa, J. FUSCA3 from barley unveils a common transcriptional regulation of seed-specific genes between cereals and Arabidopsis. Plant J. 2008, 53, 882–894. [Google Scholar] [CrossRef] [Green Version]
- Sun, F.; Liu, X.; Wie, Q.; Liu, J.; Yang, T.; Jia, L.; Wang, Y.; Yang, G.; He, G. Functional characterization of TaFUSCA3, a B3-superfamily transcription factor gene in the wheat. Front. Plant Sci. 2017, 8, 1133. [Google Scholar] [CrossRef] [Green Version]
- Guo, W.; Yang, H.; Liu, Y.; Gao, Y.; Ni, Z.; Peng, H.; Xin, M.; Hu, Z.; Sun, Q.; Yao, Y. The wheat transcription factor TaGAMyb recruits histone acetyltransferase and activates the expression of a high-molecular-weight glutenin subunit gene. Plant J. 2015, 84, 347–359. [Google Scholar] [CrossRef]
- Luo, G.; Shen, L.; Song, Y.; Yu, K.; Ji, J.; Zhang, C.; Yang, W.; Li, X.; Sun, J.; Zhan, K.; et al. The MYB family transcription factor TuODORANT1 from Triticum urartu and the homolog TaODORANT1 from Triticum aestivum inhibit seed storage protein synthesis in wheat. Plant Biotechnol. J. 2021, 19, 1863–1877. [Google Scholar] [CrossRef]
- Shen, L.; Luo, G.; Song, Y.; Xu, J.; Ji, J.; Zhang, C.; Gregová, E.; Yang, W.; Li, X.; Sun, J.; et al. A novel NAC family transcription factor SPR suppresses seed storage protein synthesis in wheat. Plant Biotechnol. J. 2021, 19, 992–1007. [Google Scholar] [CrossRef]
- Gao, Y.; An, K.; Guo, W.; Chen, Y.; Zhang, R.; Zhang, X.; Chang, S.; Rossi, V.; Jin, F.; Cao, X.; et al. The endosperm-specific transcription factor TaNAC019 regulates glutenin and starch accumulation and its elite allele improves wheat grain quality. Plant Cell 2021, 33, 603–622. [Google Scholar] [CrossRef]
- Schmidt, R.; Burr, F.A.; Aukerman, M.J.; Burr, B. Maize regulatory gene opaque-2 encodes a protein with a “leucine-zipper” motif binds to zein DNA. Proc. Natl. Acad. Sci. USA 1990, 87, 46–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravel, C.; Fiquet, S.; Boudet, J.; Dardevet, M.; Vincent, J.; Merlino, M.; Michard, R.; Martre, P. Conserved cis-regulatory modules in promoters of genes encoding wheat high-molecular-weight glutenin subunits. Front. Plant Sci. 2014, 5, 621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boudet, J.; Merlino, M.; Plessis, A.; Gaudin, J.C.; Dardevet, M.; Perrochon, S.; Alvarez, D.; Risacher, T.; Martre, P.; Ravel, C. The bZIP transcription factor SPA Heterodimerizing Protein represses glutenin synthesis in Triticum aestivum. Plant J. 2019, 97, 858–871. [Google Scholar] [CrossRef] [Green Version]
- Guo, D.; Hou, Q.; Zhang, R.; Lou, H.; Li, Y.; Zhang, Y.; You, M.; Xie, C.; Liang, R.; Li, B. Over-expressing TaSPA-B reduces prolamin and starch accumulation in wheat (Triticum aestivum L.) grains. Int. J. Mol. Sci. 2020, 21, 3257. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Somoza, I.; Martinez, M.; Diaz, I.; Carbonero, P. HvMCB1, a R1MYB transcription factor from barley with antagonistic regulatory functions during seed development and germination. Plant J. 2006, 45, 17–30. [Google Scholar] [CrossRef] [Green Version]
- Mallory, A.C.; Vaucheret, H. Functions of microRNAs and related small RNAs in plants. Nat. Genet. 2006, 38, S31–S36. [Google Scholar] [CrossRef]
- Park, W.; Li, J.; Song, R.; Messing, J.; Chen, X. Carpel factory, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002, 12, 1484–1495. [Google Scholar] [CrossRef] [Green Version]
- Chen, X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 2004, 303, 2022–2025. [Google Scholar] [CrossRef] [Green Version]
- Aukerman, M.J.; Sakai, H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef] [Green Version]
- Schmid, M.; Uhlenhaut, N.H.; Godard, F.; Demar, M.; Bressan, R.; Weigel, D.; Lohmann, J.U. Dissection of floral induction pathways using global expression analysis. Development 2003, 130, 6001–6012. [Google Scholar] [CrossRef] [Green Version]
- Schwab, R.; Palatnik, J.F.; Riester, M.; Schommer, C.; Schmid, M.; Weigel, D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell 2005, 8, 517–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debernardi, J.M.; Lin, H.Q.; Chuck, G.; Faris, J.D.; Dubcovsky, J. MicroRNA172 plays a crucial role in wheat spike morphogenesis and grain threshability. Development 2017, 144, 1966–1975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debernardi, J.M.; Greenwood, J.R.; Finnegan, E.J.; Jernstedt, J.; Dubcovsky, J. APETALA 2-like genes AP2L2 and Q specify lemma identity and axillary floral meristem development in wheat. Plant J. 2020, 101, 171–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simons, K.J.; Fellers, J.P.; Trick, H.N.; Zhang, C.; Tai, Y.S.; Gill, B.S.; Faris, J.D. Molecular characterization of the major wheat domestication gene Q. Genetics 2006, 172, 547–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.C.; Belcram, H.; Gornicki, P.; Charles, M.; Just, J.; Huneau, C.; Magdelenat, G.; Couloux, A.; Samain, S.; Gill, B.S.; et al. Duplication and partitioning in evolution and function of homoeologous Q loci governing domestication characters in polyploid wheat. Proc. Natl. Acad. Sci. USA 2011, 108, 18737–18742. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.F.; Chen, Q.; Wang, Y.; Guo, Z.R.; Xu, B.J.; Zhu, J.; Zhang, Y.Z.; Gong, X.; Luo, C.H.; Wu, W.; et al. Re-acquisition of the brittle rachis trait via a transposon insertion in domestication gene Q during wheat de-domestication. New Phytol. 2019, 224, 961–973. [Google Scholar] [CrossRef]
- Xu, B.J.; Chen, Q.; Zheng, T.; Jiang, Y.F.; Qiao, Y.Y.; Guo, Z.R.; Cao, Y.L.; Wang, Y.; Zhang, Y.Z.; Zong, L.J.; et al. An overexpressed Q allele leads to increased spike density and improved processing quality in common wheat (Triticum aestivum). G3 Genes Genom. Genet. 2018, 8, 771–778. [Google Scholar] [CrossRef] [Green Version]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Eckardt, N.A. Evolution of domesticated bread wheat. Plant Cell 2010, 22, 993. [Google Scholar] [CrossRef]
- Muramatsu, M. Dosage effect of the spelta gene q of hexaploid wheat. Genetics 1963, 48, 469–482. [Google Scholar] [CrossRef]
- Muramatsu, M. The vulgare super gene, Q: Its universality in durum wheat and its phenotypic effects in tetraploid and hexaploid wheats. Can. J. Genet. Cytol. 1986, 28, 30–41. [Google Scholar] [CrossRef]
- Endo, T.R.; Gill, B.S. The deletion stocks of common wheat. J. Hered. 1996, 87, 295–307. [Google Scholar] [CrossRef]
- Förster, S.; Schumann, E.; Baumann, M.; Weber, W.E.; Pillen, K. Copy number variation of chromosome 5A and its association with Q gene expression, morphological aberrations, and agronomic performance of winter wheat cultivars. Theor. Appl. Genet. 2013, 126, 3049–3063. [Google Scholar] [CrossRef]
- Faris, J.D.; Zhang, Q.; Chao, S.; Zhang, Z.; Xu, S. Analysis of agronomic and domestication traits in a durum × cultivated emmer wheat population using a high-density single nucleotide polymorphism-based linkage map. Theor. Appl. Genet. 2014, 127, 2333–2348. [Google Scholar] [CrossRef] [PubMed]
- Joppa, L.R.; Du, C.; Hart, G.E.; Hareland, G.A. Mapping gene (s) for grain protein in tetraploid wheat (Triticum turgidum L.) using a population of recombinant inbred chromosome lines. Crop Sci. 1997, 37, 1586–1589. [Google Scholar] [CrossRef]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubcovsky, J. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 2006, 314, 1298–1301. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Guo, Z.R.; Shi, X.L.; Wei, M.Q.; Fan, Y.Z.; Zhu, J.; Zheng, T.; Wang, Y.; Kong, L.; Deng, M.; et al. Increasing the Grain Yield and Grain Protein Content of Common Wheat (Triticum aestivum) by Introducing Missense Mutations in the Q Gene. PREPRINT (Version 1) Available at Research Square. Available online: https://www.researchsquare.com/article/rs-899571/v1 (accessed on 18 October 2021).
- Xie, Q.; Li, N.; Yang, Y.; Lv, Y.; Yao, H.; Wei, R.; Sparkes, D.L.; Ma, Z. Pleiotropic effects of the wheat domestication gene Q on yield and grain morphology. Planta 2018, 247, 1089–1098. [Google Scholar] [CrossRef]
- Mallory, A.C.; Reinhart, B.J.; Jones-Rhoades, M.W.; Tang, G.; Zamore, P.D.; Barton, M.K.; Bartel, D.P. MicroRNA control of PHABULOSA in leaf development: Importance of pairing to the microRNA 5′ region. EMBO J. 2004, 23, 3356–3364. [Google Scholar] [CrossRef] [Green Version]
- German, M.A.; Pillay, M.; Jeong, D.H.; Hetawal, A.; Luo, S.; Janardhanan, P.; Kannan, V.; Rymarquis, L.A.; Nobuta, K.; German, R.; et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 2008, 26, 941–946. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, F.; Axtell, M.J. Analysis of complementarity requirements for plant microRNA targeting using a Nicotiana benthamiana quantitative transient assay. Plant Cell 2014, 26, 741–753. [Google Scholar] [CrossRef] [Green Version]
- Chuck, G.; Meeley, R.; Irish, E.; Sakai, H.; Hake, S. The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat. Genet. 2007, 39, 1517–1521. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.H.; Upadhyaya, N.M.; Gubler, F.; Helliwell, C.A. Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa). BMC Plant Biol. 2009, 9, 149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nair, S.K.; Wang, N.; Turuspekov, Y.; Pourkheirandish, M.; Sinsuwongwat, S.; Chen, G.; Sameri, M.; Tagiri, A.; Honda, I.; Watanabe, Y.; et al. Cleistogamous flowering in barley arises from the suppression of microRNA-guided HvAP2 mRNA cleavage. Proc. Natl. Acad. Sci. USA 2010, 107, 490–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houston, K.; McKim, S.M.; Comadran, J.; Bonar, N.; Druka, I.; Uzrek, N.; Cirillo, E.; Guzy-Wrobelska, J.; Collins, N.C.; Halpin, C.; et al. Variation in the interaction between alleles of HvAPETALA2 and microRNA172 determines the density of grains on the barley inflorescence. Proc. Natl. Acad. Sci. USA 2013, 110, 16675–16680. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Z.; Reichel, M.; Deveson, I.; Wong, G.; Li, J.; Millar, A.A. Target RNA secondary structure is a major determinant of miR159 efficacy. Plant Physiol. 2017, 174, 1764–1778. [Google Scholar] [CrossRef] [Green Version]
- Chou, P.Y.; Fasman, G.D. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry 1974, 13, 211–222. [Google Scholar] [CrossRef]
- Barlow, D.J.; Thornton, J.M. Helix geometry in proteins. J. Mol. Biol. 1988, 201, 601–619. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L.; Doyle, J.A.; Doyle, F.J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Long, X.Y.; Wang, J.R.; Ouellet, T.; Rocheleau, H.; Wei, Y.M.; Pu, Z.E.; Jiang, Q.T.; Lan, X.J.; Zheng, Y.L. Genome-wide identification and evaluation of novel internal control genes for Q-PCR based transcript normalization in wheat. Plant Mol. Biol. 2010, 74, 307–311. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [Green Version]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The vienna RNA websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorenz, R.; Bernhart, S.H.; Höner zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Deleage, G.; Combet, C.; Blanchet, C.; Geourjon, C. ANTHEPROT: An integrated protein sequence analysis software with client/server capabilities. Comput. Biol. Med. 2001, 31, 259–267. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [Green Version]
- Zhao, K.; Xiao, J.; Liu, Y.; Chen, S.; Yuan, C.; Cao, A.; You, M.F.; Yang, D.; An, S.; Wang, H.; et al. Rht23 (5Dq’) likely encodes a Q homeologue with pleiotropic effects on plant height and spike compactness. Theor. Appl. Genet. 2018, 131, 1825–1834. [Google Scholar] [CrossRef]
- Zhang, Y.; Skolnick, J. TM-align: A protein structure alignment algorithm based on TM-score. Nucleic Acids Res. 2005, 33, 2302–2309. [Google Scholar] [CrossRef] [PubMed]
- Quan, L.; Lv, Q.; Zhang, Y. STRUM: Structure-based stability change prediction upon single-point mutation. Bioinformatics 2016, 32, 2936–2946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Chen, Q.; Li, Y.; Guo, Z.R.; Liu, C.H.; Wan, Y.F.; Hawkesford, M.; Zhu, J.; Wu, W.; Wei, M.Q.; et al. Post-translational cleavage of HMW-GS Dy10 allele improves the cookie-making quality in common wheat (Triticum aestivum). Mol. Breed. 2021, 41, 49. [Google Scholar] [CrossRef]
- Dupont, F.M.; Chan, R.; Lopez, R.; Vensel, W.H. Sequential extraction and quantitative recovery of gliadins, glutenins, and other proteins from small samples of wheat flour. J. Agr. Food Chem. 2005, 53, 1575–1584. [Google Scholar] [CrossRef]
- Zheng, T.; Qi, P.F.; Cao, Y.L.; Han, Y.N.; Ma, H.L.; Guo, Z.R.; Wang, Y.; Qiao, Y.Y.; Hua, S.Y.; Yu, H.Y.; et al. Mechanisms of wheat (Triticum aestivum) grain storage proteins in response to nitrogen application and its impacts on processing quality. Sci. Rep. 2018, 8, 11928. [Google Scholar] [CrossRef]
- Brandt, K.M.; Gunn, H.; Moretti, N.; Zemetra, R.S. A streamlined protocol for wheat (Triticum aestivum) protoplast isolation and transformation with CRISPR-Cas ribonucleoprotein complexes. Front. Plant Sci. 2020, 11, 769. [Google Scholar] [CrossRef]
- Song, Y.; Li, G.; Nowak, J.; Zhang, X.; Xu, D.; Yang, X.; Huang, G.; Liang, W.; Yang, L.; Wang, C.; et al. The rice actin-binding protein RMD regulates light-dependent shoot gravitropism. Plant Physiol. 2019, 181, 630–644. [Google Scholar] [CrossRef] [Green Version]
- Hellens, R.P.; Allan, A.C.; Friel, E.N.; Bolitho, K.; Grafton, K.; Templeton, M.D.; Karunairetnam, S.; Gleave, A.P.; Laing, W.A. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 2005, 1, 13. [Google Scholar] [CrossRef] [Green Version]
- Tang, Q.Y.; Zhang, C.X. Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci. 2013, 20, 254–260. [Google Scholar] [CrossRef]
- Gil-Humanes, J.; Pistón, F.; Martín, A.; Barro, F. Comparative genomic analysis and expression of the APETALA2-like genes from barley, wheat, and barley-wheat amphiploids. BMC Plant Biol. 2009, 9, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Gene ID | Log2FC | Gene Name |
|---|---|---|
| TraesCSU02G137200 | 1.42 | SPR-A [22] |
| TraesCS2B02G616900 | −3.89 | SPR-B [22] |
| TraesCS2D02G567000 | −8.22 | SPR-D [22] |
| TraesCS3A02G077900 | −0.31 | TaNAC019-A [23] |
| TraesCS3B02G092800 | −0.0037 | TaNAC019-B [23] |
| TraesCS3D02G078500 | 0.996 | TaNAC019-D [23] |
| TraesCS5A02G440400 | 0.32 | TaSHP [26] |
| TraesCS5B02G444100 | 0.46 | TaSHP [26] |
| TraesCS5D02G447500 | 0.34 | TaSHP [26] |
| TraesCS1B02G343500 | 0.13 | SPA-B [14,24] |
| TraesCS1D02G332200 | 0.35 | SPA-D [14,24] |
| TraesCS3A02G336500 | −0.61 | TaGAMYB-A [20] |
| TraesCS3B02G367500 | 0.58 | TaGAMYB-B [20] |
| TraesCS3D02G329400 | 0.49 | TaGAMYB-D [20] |
| TraesCS3A02G249100 | −0.07 | TaFUSCA3 [19] |
| TraesCS3B02G278000 | −0.38 | TaFUSCA3 [19] |
| TraesCS3D02G249100 | 0.07 | TaFUSCA3 [19] |
| TraesCS5A02G155900 | 0.47 | TaPBF |
| TraesCS5B02G154100 | 0.49 | TaPBF |
| TraesCS5D02G161000 | 0.14 | TaPBF |
| TraesCS7A02G205100 | −0.23 | TaODORANT1 [21] |
| TraesCS7B02G112400 | 0.12 | TaODORANT1 [21] |
| TraesCS7D02G208000 | −0.04 | TaODORANT1 [21] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Z.; Chen, Q.; Zhu, J.; Wang, Y.; Li, Y.; Li, Q.; Zhao, K.; Li, Y.; Tang, R.; Shi, X.; et al. The Qc5 Allele Increases Wheat Bread-Making Quality by Regulating SPA and SPR. Int. J. Mol. Sci. 2022, 23, 7581. https://doi.org/10.3390/ijms23147581
Guo Z, Chen Q, Zhu J, Wang Y, Li Y, Li Q, Zhao K, Li Y, Tang R, Shi X, et al. The Qc5 Allele Increases Wheat Bread-Making Quality by Regulating SPA and SPR. International Journal of Molecular Sciences. 2022; 23(14):7581. https://doi.org/10.3390/ijms23147581
Chicago/Turabian StyleGuo, Zhenru, Qing Chen, Jing Zhu, Yan Wang, Yang Li, Qingcheng Li, Kan Zhao, Yue Li, Rui Tang, Xiaoli Shi, and et al. 2022. "The Qc5 Allele Increases Wheat Bread-Making Quality by Regulating SPA and SPR" International Journal of Molecular Sciences 23, no. 14: 7581. https://doi.org/10.3390/ijms23147581