Discrete Typing Units of Trypanosoma cruzi Identified by Real-Time PCR in Peripheral Blood and Dejections of Triatoma infestans Used in Xenodiagnosis Descriptive Study
Abstract
:1. Introduction
2. Results
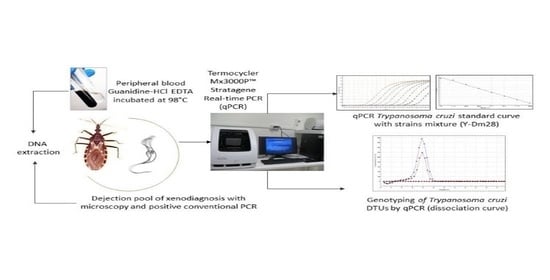
2.1. Parasite Load in Blood Samples and Dejections of XD
2.2. Genotyping in Blood Samples and Dejections of XD
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Biological Samples of Xenodiagnosis
4.3. DNA Extraction of Dejections Obtained from Xenodiagnosis (qPCR-XD)
4.4. Obtention and Purification of DNA from Blood Samples (BS) for qPCR (qPCR-BS)
4.5. PCR in Real Time (qPCR)
4.6. Trypanosoma cruzi Quantification
4.7. Genotyping by Real-Time PCR
4.8. DTU Identification of T. cruzi
4.9. Validation of the DTUs Genotyping Protocol
4.10. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Organización Mundial de la Salud (OMS). La Enfermedad de Chagas (Tripanosomiasis Americana). 2021. Available online: https://www.who.int/es/news-room/fact-sheets/detail/chagas-disease-(american-trypanosomiasis) (accessed on 16 June 2022).
- Ministerio de Salud (MINSAL). Norma General Técnica Control y Prevencioón Nacional de la Enfermedad de Chagas. Febrero. Chile. 2014. Available online: https://www.minsal.cl/sites/default/files/NORMA%20TECNICA_CHAGAS_FINAL.pdf (accessed on 16 June 2022).
- Organización Mundial de la Salud. Chagas disease, Chile: Certification of Interruption of Transmission. Wkly. Epidemiol. Rec. WER 2000, 75, 10–12. Available online: https://apps.who.int/iris/handle/10665/231033 (accessed on 16 June 2022).
- Bacigalupo, A.; Correa, J.P.; García, A.; Cattan, P.E. Focos silvestres de Triatoma infestans en Latinoamérica: Análisis y perspectivas para Chile. Parasitol. Latinoam. 2015, 64, 27–35. [Google Scholar]
- Canals, M.L.; Cifuentes, A.C.; Ayala, S.; Tapia-Garay, V.; Lillo, D.C. Eco-epidemiology of Chagas disease in Chile. In Chagas Disease: Basic Investigations and Challenges; Nissapatorn, V., Oz, H.S., Eds.; IntechOpen: London, UK, 2018; pp. 73–94. [Google Scholar]
- Zingales, B.; Andrade, S.G.; Briones, M.R.S.; Campbell, D.A.; Chiari, E.; Fernandes, O.; Gühl, F.; Lages-Silva, E.; Macedo, A.M.; Machado, C.R.; et al. A new consensus for Trypanosoma cruzi intraspecific nomenclature: Second revision meeting recommends TcI to TcVI. Mem. Inst. Oswaldo Cruz 2009, 104, 1051–1054. [Google Scholar] [CrossRef] [PubMed]
- Marcili, A.; Lima, L.; Valente, V.C.; Valente, S.A.; Batista, J.S.; Junqueira, A.C.; Souza, A.I.; da Rosa, J.A.; Campaner, M.; Lewis, M.D.; et al. Comparative phylogeography of Trypanosoma cruzi TCIIc: New hosts, association with terrestrial ecotopes, and spatial clustering. Infect. Genet. Evol. 2009, 9, 1265–1274. [Google Scholar] [CrossRef] [PubMed]
- Zingales, B. Trypanosoma cruzi genetic diversity: Something new for something known about Chagas disease manifestations, serodiagnosis and drug sensitivity. Acta Trop. 2018, 184, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Gühl, F.; Ramírez, J.D. Trypanosoma cruzi I diversity: Towards the need of genetic subdivision? Acta Trop. 2011, 119, 1–4. [Google Scholar] [CrossRef]
- Ramírez, J.D.; Duque, M.C.; Montilla, M.; Cucunubá, Z.; Guhl, F. Natural and emergent Trypanosoma cruzi I genotype revealed by mitochondrial (Cytb) and nuclear (SSU rDNA) genetic markers. Exp. Parasitol. 2012, 132, 487–494. [Google Scholar] [CrossRef] [Green Version]
- Zafra, G.; Mantilla, J.C.; Valadares, H.M.; Macedo, A.M.; González, C.I. Evidence of Trypanosoma cruzi II infection in Colombian chagasic patients. Parasitol. Res. 2008, 103, 731–734. [Google Scholar] [CrossRef]
- Dorn, P.L.; McClure, A.G.; Gallaspy, M.D.; Waleckx, E.; Woods, A.S.; Monroy, M.C.; Stevens, L. The diversity of the Chagas parasite, Trypanosoma cruzi, infecting the main Central American vector, Triatoma dimidiata, from Mexico to Colombia. PLoS Negl. Trop. Dis. 2017, 11, e0005878. [Google Scholar] [CrossRef] [Green Version]
- Campos-Soto, R.; Ortiz, S.; Cordova, I.; Bruneau, N.; Botto-Mahan, C.; Solari, A. Interactions between Trypanosoma cruzi the Chagas disease parasite and naturally infected wild Mepraia vectors of Chile. Vector Borne Zoonotic Dis. 2016, 16, 165–171. [Google Scholar] [CrossRef] [Green Version]
- Sandoval-Rodríguez, A.; Rojo, G.; López, A.; Ortiz, S.; Saavedra, M.; Botto-Mahan, C.; Cattan, P.; Solari, A. Comparing vector competence of Mepraia gajardoi and Triatoma infestans by genotyping Trypanosoma cruzi discrete typing units present in naturally infected Octodon degus. Acta Trop. 2019, 190, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Bacigalupo, A.; Segovia, V.; García, A.; Botto-Mahan, C.; Ortiz, S.; Solari, A.; Acuña-Retamar, M.; Torres-Pérez, F.; Cattan, P. Differential pattern of infection of sylvatic nymphs and domiciliary adults of Triatoma infestans with Trypanosoma cruzi genotypes in Chile. Am. J. Trop. Med. Hyg. 2012, 87, 473–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campos, R.; Acuna-Retamar, M.; Botto-Mahan, C.; Ortiz, S.; Cattan, P.E.; Solari, A. Susceptibility of Mepraia spinolai and Triatoma infestans to different Trypanosoma cruzi strains from naturally infected rodent hosts. Acta Trop. 2007, 104, 25–29. [Google Scholar] [CrossRef] [PubMed]
- De Lana, M.; da Silveira Pinto, A.; Barnabé, C.; Quesney, V.; Noël, S.; Tibayrenc, M. Trypanosoma cruzi: Compared vectorial transmissibility of three major clonal genotypes by Triatoma infestans. Exp. Parasitol. 1998, 90, 20–25. [Google Scholar] [CrossRef] [Green Version]
- Da Silveira Pinto, A.; de Lana, M.; Bastrenta, B.; Barbabé, C.; Quesney, V.; Noël, S.; Tibayrenc, M. Compared vectorial transmissibility of pure and mixed clonal genotypes of Trypanosoma cruzi in Triatoma infestans. Parasitol. Res. 1998, 84, 348–353. [Google Scholar] [CrossRef]
- Bosseno, M.; Yacsik, N.; Vargas, F.; Brenière, S. Selection of Trypanosoma cruzi clonal genotypes (Clonet 20 and 39) isolated from Bolivian Triatomines following subculture in liquid medium. Mem. Inst. Oswaldo Cruz 2000, 95, 601–607. [Google Scholar] [CrossRef] [Green Version]
- Da Silveira Pinto, A.; de Lana, M.; Britto, C.; Bastrenta, B.; Tibayrenc, M. Experimental Trypanosoma cruzi biclonal infection in Triatoma infestans: Detection of distinct clonal genotypes using kinetoplast DNA probes. Int. J. Parasitol. 2000, 30, 843–848. [Google Scholar] [CrossRef]
- Ortiz, S.; Zulantay, I.; Apt, W.; Saavedra, M.; Solari, A. Transferability of Trypanosoma cruzi from mixed human host infection to Triatoma infestans and from insect to axenic culture. Parasitol. Int. 2015, 64, 33–36. [Google Scholar] [CrossRef]
- Hernández, C.; Cucunubá, Z.; Flórez, C.; Olivera, M.; Valencia, C.; Zambrano, P.; León, C.; Ramírez, J.D. Molecular diagnosis of Chagas disease in Colombia: Parasitic loads and discrete typing units in patients from acute and chronic phases. PLoS Negl. Trop. Dis. 2016, 10, e0004997. [Google Scholar]
- Saavedra, M.; Zulantay, I.; Apt, W.; Castillo, J.; Araya, E.; Martínez, G.; Rodríguez, J. Quantification by real-time PCR of Trypanosoma cruzi DNA in samples of Triatoma infestans used in xenodiagnosis of chronic Chagas disease patients. Parasit. Vectors 2016, 9, 382. [Google Scholar] [CrossRef] [Green Version]
- Moreira, O.C.; Ramírez, J.D.; Velázquez, E.; Melo, M.F.; Lima-Ferreira, C.; Guhl, F.; Sosa-Estani, S.; Marin-Neto, J.A.; Morillo, C.A.; Britto, C. Towards the establishment of a consensus real-time qPCR to monitor Trypanosoma cruzi parasitemia in patients with chronic Chagas disease cardiomyopathy: A substudy from the BENEFIT trial. Acta Trop. 2013, 125, 23–31. [Google Scholar] [CrossRef] [PubMed]
- D’Ávila, D.A.; Galvão, L.M.C.; Sousa, G.R.; Britto, C.; Moreira, O.C.; Chiari, E. Monitoring the parasite load in chronic Chagas disease patients: Comparison between blood culture and quantitative real time PCR. PLoS ONE 2018, 13, e0208133. [Google Scholar] [CrossRef] [PubMed]
- Apt, W.; Arribada, A.; Zulantay, I.; Saavedra, M.; Araya, E.; Solari, A.; Ortiz, S.; Arriagada, K.; Rodríguez, J. Trypanosoma cruzi burden, genotypes, and clinical evaluation of Chilean patients with chronic Chagas cardiopathy. Parasitol. Res. 2015, 114, 3007–3018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz-San Martín, C.; Apt, W.; Zulantay, I. Real-time PCR strategy for the identification of Trypanosoma cruzi discrete typing units directly in chronically infected human blood. Infect. Genet. Evol. 2017, 49, 300–308. [Google Scholar] [CrossRef]
- Muñoz-San Martin, C.; Zulantay, I.; Saavedra, M.; Fuentealba, C.; Muñoz, G.; Apt, W. Discrete typing units of Trypanosoma cruzi detected by real-time PCR in Chilean patients with chronic Chagas cardiomyopathy. Acta Trop. 2018, 185, 280–284. [Google Scholar] [CrossRef]
- Cura, C.I.; Duffy, T.; Lucero, R.H.; Bisio, M.; Péneau, J.; Jimenez-Coello, M.; Calabuig, E.; Gimenez, M.J.; Valencia Ayala, E.; Kjos, S.A.; et al. Multiplex real-time PCR assay using TaqMan probes for the identification of Trypanosoma cruzi DTUs in biological and clinical samples. PLoS Negl. Trop. Dis. 2015, 9, e00033765. [Google Scholar] [CrossRef]
- Apt, W.; Carrasco, D.; Fuentealba, C.; Canals, M.; Muñoz, G.; Saavedra, M.; Castillo, J.P.; Zulantay, I. Chronic Chagas disease: Quantification of Trypanosoma cruzi in peripheral blood and dejections of Triatoma infestans fed by xenodiagnosis in patients with and without cardiopathy. Acta Trop. 2019, 200, 105167. [Google Scholar] [CrossRef]
- Coronado, X.; Zulantay, I.; Albrecht, H.; Rozas, M.; Apt, W.; Ortiz, S.; Rodríguez, J.; Sánchez, G.; Solari, A. Variation in Trypanosoma cruzi clonal composition detected in blood patients and xenodiagnosis triatomines: Implications in the molecular epidemiology of Chile. Am. J. Trop. Med. Hyg. 2006, 74, 1008–1012. [Google Scholar] [CrossRef]
- Canals, M.; Alvarado, S.; Cáceres, D.; Cattan, P.E. Twenty years of monitoring of mortality and fecundity of Triatoma infestans in the laboratory. Parasitol. Latinoam. 2016, 65, 54–60. [Google Scholar]
- Dujardin, J.P.; Steindel, M.; Chávez, T.; Machane, M.; Schofield, C. Changes in the sexual dimorphism of Triatominae in the transition from natural to artificial habitats. Mem. Inst. Oswaldo Cruz 1999, 94, 565–569. [Google Scholar] [CrossRef] [Green Version]
- Schofield, C.J.; Diotaiuti, L.; Dujardin, J.P. The process of domestication in triatominae. Mem. Inst. Oswaldo Cruz 1999, 94, 375–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catalá, S.; Dujardin, J.P. Antennal sensilla patterns indicate geographic and ecotopic variability among Triatoma infestans (Hemiptera: Reduviidae) populations. J. Med. Entomol. 2001, 38, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Sucerquia, L.J.; Triana-Chávez, O.; Jaramillo-Ocampo, N. Quantification of the genetic change in the transition of Rhodnius pallescens Barber, 1932 (Hemiptera: Reduviidae) from field to laboratory. Mem. Inst. Oswaldo Cruz 2009, 104, 871–877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrade, S.G. Morphological and behavioral characterization of Trypanosoma cruzi strains. Rev. Soc. Bras. Med. Trop. 1985, 18, 39–46. [Google Scholar]
- Schaub, G.A. Trypanosoma cruzi: Quantitative studies of development of two strains in small intestine and rectum of the vector Triatoma infestans. Exp. Parasitol. 1989, 68, 260–273. [Google Scholar] [CrossRef]
- Revollo, S.; Oury, B.; Laurent, J.-P.; Barnabé, C.; Quesney, V.; Carrière, V.; Noël, S.; Tibayrenc, M. Trypanosoma cruzi: Impact of clonal evolution of the parasite on its biological and medical properties. Exp. Parasitol. 1998, 89, 30–39. [Google Scholar] [CrossRef]
- Toledo, M.J.D.O.; Bahia, M.T.; Veloso, V.M.; Carneiro, C.M.; Machado-Coelho, G.L.L.; Alves, C.F.; Martins, H.R.; Cruz, R.E.; Tafuri, W.L.; Lana, M.D. Effects of specific treatment on parasitological and histopathological parameters in mice infected with different Trypanosoma cruzi clonal genotypes. J. Antimicrob. Chemother. 2004, 53, 1045–1053. [Google Scholar] [CrossRef] [Green Version]
- Margioto, A.P.; de Abreu, A.P.; Gruendling, A.P.; Bahia, M.T.; Gomes, M.L.; Marqués de Araújo, S.; Ornelas, J.M. Differential parasitological, molecular, and serological detection of Trypanosoma cruzi I, II, and IV in blood of experimentally infected mice. Exp. Parasitol. 2016, 166, 44–50. [Google Scholar] [CrossRef]
- Martins, H.R.; Toledo, M.J.; Veloso, V.M.; Carneiro, C.M.; Machado-Coelho, G.L.; Tafuri, W.L.; Bahia, M.T.; Valadares, H.M.; Macedo, A.M.; Lana, M. Trypanosoma cruzi: Impact of dual-clone infections on parasite biological properties in BALB/c mice. Exp. Parasitol. 2006, 112, 237–246. [Google Scholar] [CrossRef] [Green Version]
- Perlowagora-Szumlewicz, A.; Muller, C.A.; de Carvalho Moreira, C.J. Studies in search of a suitable experimental insect model for xenodiagnosis of hosts with Chagas’ disease. 3, On the interaction of vector species and parasite strains in the reaction of bugs to infection by Trypanosoma cruzi. Rev. Saúde Pública 1988, 22, 390–400. [Google Scholar] [CrossRef] [Green Version]
- Perlowagora-Szumlewicz, A.; Muller, C.A.; de Carvalho Moreira, C.J. Studies in search of a suitable experimental insect model for xenodiagnosis of hosts with Chagas’ disease. 4. The reflection of parasite stock in the responsiveness of different vector species to chronic infection with different Trypanosoma cruzi stocks. Rev. Saúde Pública 1990, 24, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Schijman, A.G. Molecular diagnosis of Trypanosoma cruzi. Acta Trop. 2018, 184, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Cura, C.I.; Mejía-Jaramillo, A.M.; Duffy, T.; Burgos, J.M.; Rodriguero, M.; Cardinal, M.V.; Kjos, S.; Gurgel-Gonçalves, R.; Blanchet, D.; de Pablos, L.M.; et al. Trypanosoma cruzi I genotype in different geographic regions and transmission cycles based on a microsatellite motif of the intergenic spacer of spliced leader genes. Int. J. Parasitol. 2010, 40, 1599–1607. [Google Scholar] [CrossRef] [Green Version]
- Gaunt, M.; Miles, M. The ecotopes and evolution of triatomine bugs (Triatominae) and their associated trypanosomes. Mem. Inst. Oswaldo Cruz 2000, 95, 557–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zingales, B.; Miles, M.A.; Campbell, D.A.; Tibayrenc, M.; Macedo, A.M.; Teixeira, M.M.; Schijman, A.G.; Llewellyn, M.S.; Lages-Silva, E.; Machado, C.R.; et al. The revised Trypanosoma cruzi subspecific nomenclature: Rationale, epidemiological relevance and research applications. Infect. Genet. Evol. 2012, 12, 240–253. [Google Scholar] [CrossRef] [PubMed]
- Ihle-Soto, C.; Costoya, E.; Correa, J.P.; Bacigalupo, A.; Cornejo-Villar, B.; Estadella, V.; Solari, A.; Ortiz, S.; Hernández, H.J.; Botto-Mahan, C.; et al. Spatio-temporal characterization of Trypanosoma cruzi infection and discrete typing units infecting hosts and vectors from nondomestic foci of Chile. PLoS Negl. Trop. Dis. 2019, 13, e0007170. [Google Scholar] [CrossRef] [PubMed]
- Correa, J.; Bacigalupo, A.; Yefi-Quinteros, E.; Rojo, G.; Solari, A.; Cattan, P.; Botto-Mahan, C. Trypanosomatid Infections among Vertebrates of Chile: A Systematic Review. Pathogens 2020, 9, 661. [Google Scholar] [CrossRef]
- Botto-Mahan, C.; Cattan, P.E.; Canals, M.; Acuña, M. Seasonal variation in the home range and host availability of the blood-sucking insect Mepraia spinolai in wild environment. Acta Trop. 2005, 95, 160–163. [Google Scholar] [CrossRef]
- Rozas, M.; Botto-Mahan, C.; Coronado, X.; Ortiz, S.; Cattan, P.E.; Solari, A. Coexistence of Trypanosoma cruzi genotypes in wild and peridomestic mammals in Chile. Am. J. Trop. Med. Hyg. 2007, 77, 647–653. [Google Scholar] [CrossRef]
- Schmid-Hempel, P. Host-parasite co-evolution. In Evolutionary Parasitology: The Integrated Study of Infections, Immunology, Ecology, and Genetics; Oxford University Press: Oxford, UK, 2021; pp. 241–281. [Google Scholar]
- Schofield, C.J. The biosystematics of Triatominae. In Biosystematics of Haematophagous Insects; Service, M.W., Ed.; Systematics Association/Clarendon Press: Oxford, UK, 1988; Special Volume 37, pp. 284–312. [Google Scholar]
- Panzera, F.; Ferreiro, M.J.; Pita, S.; Calleros, L.; Pérez, R.; Basmadjián, Y.; Guevara, Y.; Brenière, S.F.; Panzera, Y. Evolutionary and dispersal history of Triatoma infestans, main vector of Chagas disease, by chromosomal markers. Infect. Genet. Evol. 2014, 27, 105–113. [Google Scholar] [CrossRef]
- Schofield, C.J. Biosystematics and evolution of the Triatominae. Cad. Saúde Pública 2000, 16, S89–S92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carcavallo, R.U. Phylogeny of triatominae. The Triatoma infestans complex. Mem. Inst. Oswaldo Cruz 1998, 93, 68–70. [Google Scholar]
- Brenière, S.F.; Waleckx, E.; Barnabe, C. Over six thousand Trypanosoma cruzi strains classified into discrete typing units (DTUs): Attempt at an inventory. PLoS Negl. Trop. Dis. 2016, 10, e0004792. [Google Scholar] [CrossRef] [PubMed]
- Westenberger, S.J.; Barnabé, C.; Campbell, D.A.; Sturm, N.R. Two hybridization events define the population structure of Trypanosoma cruzi. Genetics 2005, 171, 527–543. [Google Scholar] [CrossRef] [Green Version]
- Tomasini, N.; Diosque, P. Evolution of Trypanosoma cruzi: Clarifying hybridizations, mitochondrial introgressions and phylogenetic relationships between major lineages. Mem. Inst. Oswaldo Cruz 2015, 110, 403–413. [Google Scholar] [CrossRef] [Green Version]
- Lewis, M.D.; Llewellyn, M.S.; Yeo, M.; Acosta, N.; Gaunt, M.W.; Miles, M.A. Recent, independent and anthropogenic origins of Trypanosoma cruzi hybrids. PLoS Negl. Trop. Dis. 2011, 5, e1363. [Google Scholar] [CrossRef] [Green Version]
- Usinger, R.L.; Wygodzinsky, P.; Ryckman, R.E. The biosystematics of Triatominae. Annu. Rev. Entomol. 1966, 1, 309–333. [Google Scholar] [CrossRef]
- Muñoz, G.; Vergara, C.; Martínez, G.; Apt, W.; Zulantay, I. Quantification of Immunoglobulin G against Trypanosoma cruzi in individuals with chronic Chagas disease treated with nifurtimox and evaluated in prolonged follow-up. Korean J. Parasitol. 2019, 57, 39–41. [Google Scholar] [CrossRef] [Green Version]
- Schenone, H. Xenodiagnosis. Mem. Inst. Oswaldo Cruz 1999, 94, 289–294. [Google Scholar] [CrossRef] [Green Version]
- Saavedra, M.; Bacigalupo, A.; Barrera, M.V.; Vergara, M.J.; Álvarez-Duhart, B.; Muñoz-San Martín, C.; Solís, R.; Cattan, P.E. Trypanosoma cruzi infection in the wild Chagas disease vector, Mepraia spinolai: Parasitic load, discrete typing units, and blood meal sources. Acta Trop. 2022, 229, 106365. [Google Scholar] [CrossRef]
| Patient | Sex | Age | Xenodiagnosis | Peripheral Blood | ||
|---|---|---|---|---|---|---|
| par. eq/mL | DTU | par. eq/mL | DTU | |||
| 1 | M | 32 | 140,650 | TcII | 4.79 | - |
| 2 | M | 58 | 101.5 | TcV | 6.88 | - |
| 3 | F | 53 | 12.65 | - | 22.4 | TcV |
| 4 | F | 53 | 1.79 | - | 31.1 | TcI |
| 5 | F | 33 | 221 | TcV | 17.6 | - |
| 6 | F | 63 | 51.5 | TcV | 26.75 | - |
| 7 | M | 54 | 134 | TcI-TcV | 4.76 | - |
| 8 | M | 68 | 51 | TcV | 13 | TcV |
| 9 | M | 76 | 1388.50 | TcII-TcV | 90.4 | TcV |
| 10 | F | 20 | 653.5 | TcII-TcV | 9.46 | TcII-TcV |
| 11 | F | 53 | 160.5 | TcV | 4.24 | - |
| 12 | M | 56 | 3277 | TcI-TcV | 1.55 | - |
| 13 | M | 58 | 9.37 | TcII-TcV | 0.72 | - |
| 14 | F | 53 | 486.5 | TcV | 17.2 | - |
| Trypanosoma cruzi DTU | Xenodiagnosis % | Peripheral Blood Samples % |
|---|---|---|
| TcI | 0 | 7.14 |
| TcII | 7.14 | 0 |
| TcV | 42.85 | 21.42 |
| TcI + TcV | 14.28 | 0 |
| TcII + TcV | 21.42 | 7.14 |
| Undetected DTU | 14.28 | 64.28% |
| Patient | Sex | Age | Xenodiagnosis | Location | ||
|---|---|---|---|---|---|---|
| 30 Days | 60 Days | 90 Days | ||||
| 1 | M | 32 | (−) | (+) | ND | Illapel |
| 2 | M | 58 | (−) | (+) | (−) | Combarbalá |
| 3 | F | 53 | (−) | (−) | (+) | Combarbalá |
| 4 | F | 53 | (+) | (−) | (+) | Salamanca |
| 5 | F | 33 | (−) | (+) | (+) | Combarbalá |
| 6 | F | 63 | (−) | (+) | (+) | Combarbalá |
| 7 | M | 54 | (−) | 2 (+) | 2 (+) | Combarbalá |
| 8 | M | 68 | (−) | 2 (+) | 2 (+) | Combarbalá |
| 9 | M | 76 | 2 (+) | 2 (+) | 2 (+) | Combarbalá |
| 10 | F | 20 | (−) | (+) | (+) | Salamanca |
| 11 | F | 53 | (−) | 2 (+) | 2 (+) | Salamanca |
| 12 | M | 56 | (+) | 2 (+) | 2 (+) | Illapel |
| 13 | M | 58 | (+) | (+) | (+) | Illapel |
| 14 | F | 53 | 2 (+) | (+) | (+) | Salamanca |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zulantay, I.; Muñoz, G.; Liempi, D.; Rozas, T.; Manneschi, M.J.; Muñoz-San Martín, C.; Botto-Mahan, C.; Apt, W.; Cabrera, G. Discrete Typing Units of Trypanosoma cruzi Identified by Real-Time PCR in Peripheral Blood and Dejections of Triatoma infestans Used in Xenodiagnosis Descriptive Study. Pathogens 2022, 11, 787. https://doi.org/10.3390/pathogens11070787
Zulantay I, Muñoz G, Liempi D, Rozas T, Manneschi MJ, Muñoz-San Martín C, Botto-Mahan C, Apt W, Cabrera G. Discrete Typing Units of Trypanosoma cruzi Identified by Real-Time PCR in Peripheral Blood and Dejections of Triatoma infestans Used in Xenodiagnosis Descriptive Study. Pathogens. 2022; 11(7):787. https://doi.org/10.3390/pathogens11070787
Chicago/Turabian StyleZulantay, Inés, Gabriela Muñoz, Daniela Liempi, Tamara Rozas, María José Manneschi, Catalina Muñoz-San Martín, Carezza Botto-Mahan, Werner Apt, and Gonzalo Cabrera. 2022. "Discrete Typing Units of Trypanosoma cruzi Identified by Real-Time PCR in Peripheral Blood and Dejections of Triatoma infestans Used in Xenodiagnosis Descriptive Study" Pathogens 11, no. 7: 787. https://doi.org/10.3390/pathogens11070787