Scanning Electron Microscopy and First Molecular Data of Two Species of Lamproglena (Copepoda: Lernaeidae) from Labeo victorianus (Cyprinidae) and Clarias gariepinus (Clariidae) in Kenya
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection, Examination, and Identification
2.2. Morphological Analyses
2.3. DNA Extraction, PCR, and Sequencing
2.4. Phylogenetic Analyses
3. Results
3.1. Taxonomic Summary
- Lamproglena cleopatra Humes, 1957.
- Host: Labeo victorianus Boulenger, 1901 (Cypriniformes: Cyprinidae).
- Site of infection: Gills.
- Locality/collection date: Nyando River-Ahero (Lake Victoria drainage system), KisumuCounty, Kenya (0°0′ 0°22′ S, 34°51′ E 35°11′ E), collected 10 May 2022 and 10 March 2023 by Drs. Nehemiah M. Rindoria and George N. Morara.
- Materials studied: 14 specimens (5 for morphometrics, 4 for SEM, and 5 for molecular analysis).
- Deposition of voucher specimens: A total of six voucher female specimens were deposited in the Helminthological Collection of the Institute of Parasitology, the Biology Centre of the Czech Academy of Sciences, České Budějovice, Czech Republic (IPCAS Cr-38).
- Deposition of sequences: Sequence data obtained were deposited in GenBank: 18S rDNA (OR242501, OR242502), 28S rDNA (OR338169, OR338170), and cox1 (OR232207).
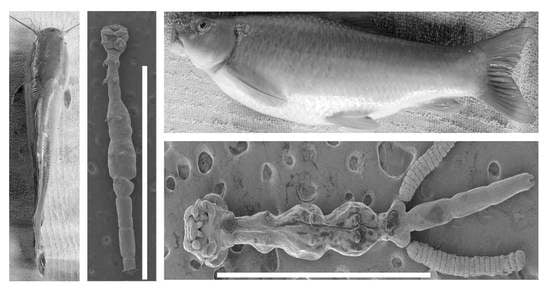
- Redescription (Figure 1) Female (based on nine specimens, five morphometrics (all measurements in millimetres), and four SEM): Body elongated, slender, cylindrical, total length (excluding caudal rami) 2.71 (2.41–3.20) (Figure 1A,B). Body divided into cephalothorax, thorax, and abdomen (Figure 1A,B). Cephalothorax length 0.43 (0.36–0.54), width 0.56 (0.51–0.62), width represents 20.20% of total length, laterally indented; wider posterior part than thorax; U-shaped ridge on dorsal surface (Figure 1A,B). First thoracic segment fused with the head (Figure 1A–D). Second, third, and fourth thoracic segments free, with pedigerous segments distinct and well separated with indentations laterally (Figure 1A,B). Second segment 0.35 (0.24–0.42) wide, 0.26 (0.19–0.31) long. Third and fourth segments 0.42 (0.35–0.53) and 0.50 (0.37–0.54) long, respectively; width subequal 0.51 (0.39–0.59) and 0.50 (0.41–0.56), respectively, wider than the second segment (Figure 1A,B). Fifth thoracic segment narrower 0.24 (0.15–0.27), shorter 0.096 (0.07–0.13), bearing tiny fifth legs (Figure 1A,B,K). Genital segment free, wider 0.354 (0.31–0.40) than fifth thoracic segment, 0.19 (0.13–0.24) long, with egg sacs attached laterally (Figure 1A); other specimens with chitinous, kidney-shaped spermatophores attached ventrally (Figure 1A,B,L). Abdomen length 0.94 (0.79–1.10) (about 34.23% (29.43–37.69) of the total body length) composed of three approximately equal, poorly demarcated segments (Figure 1A,B). Furcal rami (Figure 1A,B,M,N) minute, 0.028 (0.02–0.03) wide, 0.037 (0.03–0.04) long. Each ramus with one long seta, one pore on inner and outer margins, and terminally with four setae, one blunt process, and two pores (Figure 1N). Antennules uniramous, indistinctly two-jointed with long swollen basal podomere bearing 11–14 naked setae and small distal podomere with 5 naked setae, 1 lateral and 4 terminal. Dorsal side of antennule with circular pores (Figure 1C–E). Antenna uniramous, indistinctly four-jointed, distal segment with five small terminal setae (Figure 1C–E). Oral region consisting of distinct projecting sucker-like with two lateral lobes from which arises two long setae and two finger-like posterior lobes (Figure 1C–E). Mandible not observed. Maxilla uniramous, rigid, covered with a thin layer through which distinct terminal spine projects, basal region finely granulated (Figure 1A–E). Maxilliped equipped with three roughly equal, curved claws, with a minute spine-like protrusion on the proximal part (Figure 1F). Legs 1–4 biramous, rami of legs indistinctly two-jointed. Endopodites of legs 1–4 all similar, terminating in a minute, rather blunt seta. Protopodite of legs 1–4 with one lateral long seta at the base before exopodite (Figure 1G–J). Exopodite of first leg first podomere with one smaller seta and four long terminal setae on the second podomere (Figure 1G). Second leg first exopodite podomere with one basal seta, second exopodite podomere with two small setae and a minute knob, an opening between setae and knob (Figure 1H). Second exopodite podomere of third and fourth legs with four setae: two long, one medium, one min (Figure 1I,J). Fifth leg made of small lobe with two long distal and one lateral seta (Figure 1K). Spermatophore observed (Figure 1I,L). Egg sac 0.98 × 0.24, containing about 20 eggs (19–22) (Figure 1A).
- Lamproglena clariae Fryer, 1956 (Figure 2).
- Host: Clarias gariepinus (Burchell, 1822) (Siluriformes, Clariidae).
- Site of infection: Gills.
- Locality/collection date: Nyando River-Ahero (Lake Victoria drainage system), Kisumu County, Kenya (0°0′ 0°22′S, 34°51′E 35°11′E), collected 10 May 2022 and 10 March 2023 by Drs. Nehemiah M. Rindoria and George N. Morara.
- Materials examined: Two specimens, one for SEM and one for molecular analysis.
- Deposition of voucher specimens: Not deposited.
- Deposition of sequences: Sequence data obtained were deposited in GenBank: 18S rRNA (OR242503, OR242504), 28S rRNA (OR338195, OR338196), and cox1 (OR232208, OR232209).
- Remarks: Based on the morphological data available from the reports of Fryer [5] and Marx and Avenant-Oldewage [14], the present material was identical to L. clariae. Following a detailed redescription of this parasite using LM and SEM by Marx and Avenant-Oldewage [14], the present study only provided the SEM images to confirm the identity of our specimen and most importantly provided genetic sequences using 18S, 28S, and cox1 markers.
3.2. Molecular Identification
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jirsa, F.; Zitek, A.; Schachner, O. First record of Lamproglena pulchella Nordmann, 1832 in the Pielach and Melk rivers, Austria. J. Appl. Ichthyol. 2006, 22, 404–406. [Google Scholar] [CrossRef]
- Kunutu, K.D.; Tavakol, S.; Halajian, A.; Baker, C.; Paoletti, M.; Heckmann, R.A.; Luus-Powell, W.J. Expanded description of Lamproglena cleopatra Humes, 1957 (Lernaeidae: Copepoda) from Labeo spp. (Cyprinidae) with a key to species of Lamproglena von Nordmann, 1832. Syst. Parasitol. 2018, 95, 91–103. [Google Scholar] [CrossRef]
- Piasecki, W. Comparative morphology of the three species of Lamproglena (Copepoda, Cyclopoida, Lernaeidae) described by von Nordmann, based on re-examination of the types. Mitt. Zool. Mus. Berl. 1993, 69, 307–315. [Google Scholar] [CrossRef]
- Capart, A. Notes sur les Copépodes parasites. III. Copépodes parasites des poissons d’eau douce du Congo Belge. Bull. Mus. R. Hist. Nat. Belg. 1944, 20, 1–24. [Google Scholar]
- Fryer, G. A report on the parasitic Copepoda and Branchiura of the fishes of Lake Nyasa. Proc. Zool. Soc. Lond. 1956, 127, 29–44. [Google Scholar] [CrossRef]
- Fryer, G. The parasitic Copepoda and Branchiura of the fishes of Lake Victoria and the Victoria Nile. Proc. Zool. Soc. Lond. 1961, 137, 41–60. [Google Scholar] [CrossRef]
- Rindoria, N.M.; Dos Santos, Q.M.; Ali, S.E.; Ibraheem, M.H.; Avenant Oldewage, A. Lamproglena monodi Capart, 1944 infecting Oreochromis niloticus (Linnaeus, 1758): Additional information on infection, morphology and genetic data. Afr. Zool. 2022, 57, 98–110. [Google Scholar] [CrossRef]
- Humes, A.G. Two new caligoid copepods from Egyptian fishes. J. Parasitol. 1957, 43, 201–208. [Google Scholar] [CrossRef]
- Fryer, G. Further studies on the parasitic Crustacea of African freshwater fishes. Proc. Zool. Soc. Lond. 1964, 143, 79–102. [Google Scholar]
- Fryer, G. The parasitic Crustacea of African freshwater fishes; their biology and distribution. J. Zool. 1968, 156, 45–95. [Google Scholar] [CrossRef]
- Thurston, P. The incidence of Monogenea and parasitic Crustacea on the gills of fish in Uganda. Rev. Zool. Bot. Afric. 1970, 82, 111–130. [Google Scholar]
- Shotter, R.A. Copepod parasites of fishes from northern Nigeria. Bull. I.F.A.N. 1977, 39, 583–600. [Google Scholar]
- Uler, C.; Avenant-Oldewage, A. Die biologie van verteenwoordiges van die genus Lamproglena. Suid-Afrik. Tydskr. Nat. Wet. Tegnol. 1992, 11, 72–73. [Google Scholar]
- Marx, H.M.; Avenant-Oldewage, A. Redescription of Lamproglena clariae Fryer, 1956 (Copepoda: Lernaeidae) with notes on its occurrence and distribution. Crustaceana 1996, 69, 509–523. [Google Scholar]
- Rindoria, N.M.; Moraa, G.N.; Smit, W.J.; Truter, M.; Smit, N.J.; Luu-Powell, W.J. Integrated morphological and molecular characterization of the fish parasitic nematode Rhabdochona (Rhabdochona) gendrei Campana-Rouget, 1961 infecting Labeobarbus altianalis (Boulenger, 1900) in Kenya. IJP-PAW 2023, 21, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Okeyo, D.O.; Ojwang, W.O. A Photographic Guide to Freshwater Fishes of Kenya; 2015; p. 66. Available online: www.seriouslyfish.com/publications/ (accessed on 1 March 2022).
- Froese, R.; Pauly, D. (Eds.) FishBase. World Wide Web Electronic Publication; 2013; Available online: www.fishbase.org (accessed on 3 August 2022).
- Schäperclaus, W. Fischkrankheiten; Akademie Verlag: Berlin, Germany, 1990. [Google Scholar]
- Boxshall, G.A.; Halsey, S.H. An Introduction to Copepod Diversity; Part II; Ray Society: London, UK, 2004. [Google Scholar]
- Nation, J.L. A new method using hexamethyldisilazane for preparation of soft insect tissues for scanning electron microscopy. Stain Technol. 1983, 58, 347–351. [Google Scholar] [CrossRef]
- Dos Santos, Q.M.; Avenant-Oldewage, A. Soft tissue digestion of Paradiplozoon vaalense for SEM of sclerites and simultaneous molecular analysis. J. Parasitol. 2015, 101, 94–97. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Song, Y.; Wang, G.T.; Yao, W.J.; Gao, Q.; Nei, P. Phylogeny of freshwater parasitic copepods in the Ergasilidae (Copepoda: Poecilostomatoida) based on 18S and 28S rDNA sequences. Parasitol. Res. 2008, 102, 299–306. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods. 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Rambaut, A. FigTree v. 1.4. In Molecular Evolution, Phylogenetics and Epidemiology; University of Edinburgh, Institute of Evolutionary Biology: Edinburgh, UK, 2012; Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 1 August 2022).
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Mabika, N.; Barson, M.; Dos Santos, Q.M.; van Dyk, C.; Avenant-Oldewage, A. Additional Taxonomic Information for Lamproglena hemprichii (Copepoda: Lernaeidae) Based on Scanning Electron Microscopy and Genetic Characterization, Alongside Some Aspects of its Ecology. Zool. Sci. 2023, 40. [Google Scholar] [CrossRef]




| Humes [8] | Kunutu et al. [2] | Present Study | ||
| Country/fish species/no. measured | Egypt: L. forskalii n = 5 | SA: L. rosae and L. molybdinus ZIM: L. ruddi n = 40 | KEN: L. victorianus n = 5 | |
| Taxonomic feature | ||||
| Total length | 2.60 (2.43–2.77) | 2.79 ± 0,39 (1.66–3.38) | 2.71 ± 0.30 (2.41–3.20) | |
| Cephalothorax | L | 0.504 | - | 0.43 ± 0.07 (0.36–0.54) |
| W | 0.375 | 0.58 ± 0.07 (0.41–0.71) | 0.56 ± 0.05 (0.51–0.62) | |
| Second thoracic segment | L | - | 0.28 ± 0.07 (0.16–0.41) | 0.26 ± 0.05 (0.19–0.31) |
| W | 0.291 | 0.32 ± 0.05 (0.19–0.40) | 0.35 ± 0.07 (0.24–0.42) | |
| Third thoracic segment | L | - | 0.38 ± 0.06 (0.15–0.48) | 0.42 ± 0.07 (0.35–0.53) |
| W | 0.422 | 0.43 ± 0.08 (0.20–0.59) | 0.52 ± 0.08 (0.39–0.59) | |
| Fourth thoracic segment | L | - | 0.41 ± 0.07 (0.16–0.51) | 0.50 ± 0.07 (0.37–0.54 |
| W | 0.413 | 0.43 ± 0.08 (0.20–0.59 | 0.50 ± 0.06 (0.41–0.56) | |
| Fifth leg-bearing segment | L | - | 0.09 ± 0.02 (0.06–0.14) | 0.096 ± 0.02 (0.07–0.13) |
| W | 0.212 | 0.22 ± 0.03 (0.16–0.30) | 0.242 ± 0.05 (0.15–0.29) | |
| Genital segment | L | - | 0.17 ± 0.03 (0.13–0.22) | 0.194 ± 0.04 (0.13–0.24) |
| W | 0.343 | 0.35 ± 0.06 (0.16–0.43) | 0.354 ± 0.02 (0.31–0.40) | |
| Egg sac | L | 1.32 | 1.22 ± 0.23 (0.92–1.46) | 0.976 (n = 1) |
| W | 0.171 | - | 0.24 (n = 1) | |
| Abdomen | L | 0.975 | 0.96 ± 0.16 (0.56–1.22) | 0.94 ± 0.13 (0.79–1.10) |
| W | - | 0.19 ± 0.02 (0.14–0.25) | - | |
| % of the abdomen to total body length | 37 | 34 | 34 | |
| Furcal rami | L | 0.039 | 0.04 ± 0.01 (0.03–0.06) | 0.037 (0.03–0.04) |
| W | 0.026 | 0.028 (0.02–0.03) | ||
| Species | Host | Family | Locality | 18S | 28S | cox1 | Reference |
|---|---|---|---|---|---|---|---|
| Lamproglena orientalis | Squaliobarbus curriculus | Xenocyprididae | Dangjiangkou Reservoir, China | DQ107552 | DQ107544 | ― | Song et al. [2] |
| Lamproglena orientalis | Chanodichthys erythropterus | Xenocyprididae | Tangxun Lake, China | DQ107551 | DQ107541 | Song et al. [2] | |
| Lamproglena orientalis | Chanodichthys mongolicus | Xenocyprididae | E-zhou farm, China | DQ107550 | DQ107543 | ― | Song et al. [2] |
| Lamproglena orientalis | Chanodichthys dabryi | Xenocyprididae | Tangxun Lake, China | DQ107549 | DQ107542 | ― | Song et al. [2] |
| Lamproglena hemprichii | Hydrocynus vittatus | Alestidae | Lake Kariba, Zimbabwe | OP277526 | OP277527 | ― | Mabika et al. [28] |
| Lamproglena cleopatra Isolate UL236 | Labeo victorianus | Cyprinidae | Nyando River, Kenya | OR242501 | OR338169 | ― | Present study |
| Lamproglena cleopatra Isolate UL237 | Labeo victorianus | Cyprinidae | Nyando River, Kenya | OR242502 | OR338170 | OR232207 | Present study |
| Lamproglena clariae Isolate UL241 | Clarias gariepinus | Clariidae | Nyando River, Kenya | OR242503 | OR338195 OR338196 | OR232208 | Present study |
| Lamproglena clariae Isolate UL242 | Clarias gariepinus | Clariidae | Nyando River, Kenya | OR242504 | ― | OR232209 | Present study |
| Lamproglena monodi | Oreochromis niloticus | Cichlidae | Kibos Fish Farm, Kenya | ON419439 | ON419422 | ― | Rindoria et al. [7] |
| Lamproglena monodi | Oreochromis niloticus | Cichlidae | Kibos Fish Farm, Kenya | ON419444 | ON419428 | ― | Rindoria et al. [7] |
| Lamproglena monodi | Oreochromis niloticus | Cichlidae | Sharqia, Egypt | ON419450 | ON419435 | ― | Rindoria et al. [7] |
| Lamproglena monodi | Oreochromis niloticus | Cichlidae | El-Minia, Egypt | ON419448 | ON419432 | ― | Rindoria et al. [7] |
| Lamproglena chinensis | Channa argus | Channidae | Dangjiangkou Reservoir | DQ107553 | DQ107545 | ― | Song et al. [2] |
| Lernea cyprinacea | Chanodichthys erythropterus | Xenocyprididae | Lake Dongxi, China | DQ107556 | DQ107548 | ― | Song et al. [2] |
| Accession Number | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | L. cleopatra UL236 | OR242501 | 0.0 | 1.0 | 1.0 | 0.1 | 0.2 | 0.2 | 0.2 | 0.2 | 1.2 | 1.2 | 1.4 | 2.0 | 2.3 | |
| 2 | L. cleopatra UL237 | OR242502 | 0 | 1.0 | 1.0 | 0.1 | 0.2 | 0.2 | 0.2 | 0.2 | 1.2 | 1.2 | 1.4 | 2.0 | 2.3 | |
| 3 | L. clariae UL241 | OR242503 | 14 | 14 | 0.0 | 0.9 | 0.9 | 0.9 | 0.9 | 0.9 | 1.7 | 1.7 | 1.9 | 2.0 | 2.4 | |
| 4 | L. clariae UL242 | OR242504 | 14 | 14 | 0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.9 | 1.9 | 1.9 | 2.1 | 2.6 | |
| 5 | L. hemprichii | OP277526 | 1 | 1 | 13 | 13 | 0.3 | 0.3 | 0.3 | 0.3 | 1.1 | 1.1 | 1.4 | 2.0 | 2.4 | |
| 6 | L. monodi | ON419439 | 3 | 3 | 13 | 13 | 4 | 0.0 | 0.0 | 0.0 | 1.3 | 1.3 | 1.5 | 2.0 | 2.4 | |
| 7 | L. monodi | ON419444 | 3 | 3 | 13 | 13 | 4 | 0 | 0.0 | 0.0 | 1.3 | 1.3 | 1.5 | 2.0 | 2.4 | |
| 8 | L. monodi | ON419448 | 3 | 3 | 13 | 13 | 4 | 0 | 0 | 0.0 | 1.3 | 1.3 | 1.5 | 2.0 | 2.4 | |
| 9 | L. monodi | ON419450 | 3 | 3 | 13 | 13 | 4 | 0 | 0 | 0 | 1.3 | 1.3 | 1.5 | 2.0 | 2.4 | |
| 10 | L. orientalis | DQ107549 | 16 | 16 | 24 | 24 | 15 | 17 | 17 | 17 | 17 | 0.0 | 0.3 | 2.2 | 2.6 | |
| 11 | L. orientalis | DQ107550 | 16 | 16 | 24 | 24 | 15 | 17 | 17 | 17 | 17 | 0 | 0.3 | 2.2 | 2.6 | |
| 12 | L. orientalis | DQ107552 | 19 | 19 | 26 | 25 | 19 | 20 | 20 | 20 | 20 | 4 | 4 | 2.4 | 2.8 | |
| 13 | L. chinensis | DQ107553 | 30 | 30 | 29 | 28 | 29 | 29 | 29 | 29 | 29 | 32 | 32 | 35 | 2.5 | |
| 14 | Lernaea cyprinacea | DQ107556 | 33 | 33 | 35 | 35 | 34 | 34 | 34 | 34 | 34 | 37 | 37 | 39 | 38 |
| Accession Number | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | L. cleopatra UL236 | OR338169 | 0.0 | 19.2 | 19.4 | 7.7 | 9.9 | 9.9 | 9.9 | 10.1 | 20.8 | 21.0 | 21.2 | 20.9 | 21.7 | 22.2 | |
| 2 | L. cleopatra UL237 | OR338170 | 0 | 20.4 | 20.4 | 7.1 | 9.1 | 9.1 | 9.1 | 9.2 | 22.4 | 22.6 | 22.7 | 22.5 | 23.3 | 23.0 | |
| 3 | L. clariae UL241 c3 | OR338195 | 115 | 135 | 1.3 | 18.4 | 16.8 | 16.8 | 16.8 | 16.9 | 23.5 | 23.7 | 23.5 | 23.3 | 21.1 | 24.0 | |
| 4 | L. clariae UL242 c25 | OR338196 | 116 | 135 | 9 | 17.9 | 16.8 | 16.8 | 16.8 | 16.9 | 23.2 | 23.4 | 23.2 | 23.0 | 20.7 | 24.0 | |
| 5 | L. hemprichii | OP277527 | 46 | 47 | 131 | 128 | 6.6 | 6.6 | 6.6 | 6.8 | 19.9 | 20.0 | 20.1 | 19.9 | 19.9 | 22.5 | |
| 6 | L. monodi | ON419422 | 59 | 60 | 120 | 120 | 48 | 0.0 | 0.0 | 0.0 | 18.7 | 18.8 | 19.0 | 18.9 | 19.4 | 22.4 | |
| 7 | L. monodi | ON419428 | 59 | 60 | 120 | 120 | 48 | 0 | 0.0 | 0.0 | 18.7 | 18.8 | 19.0 | 18.9 | 19.4 | 22.4 | |
| 8 | L. monodi | ON419432 | 59 | 60 | 120 | 120 | 48 | 0 | 0 | 0.0 | 18.7 | 18.8 | 19.0 | 18.9 | 19.4 | 22.4 | |
| 9 | L. monodi | ON419435 | 59 | 60 | 120 | 120 | 48 | 1 | 1 | 1 | 18.7 | 18.8 | 19.0 | 18.9 | 19.4 | 22.5 | |
| 10 | L. orientalis | DQ107541 | 122 | 146 | 166 | 164 | 139 | 131 | 131 | 131 | 130 | 0.1 | 0.3 | 2.5 | 21.0 | 22.2 | |
| 11 | L. orientalis | DQ107543 | 123 | 147 | 167 | 165 | 140 | 132 | 132 | 132 | 131 | 1 | 0.4 | 2.6 | 21.2 | 22.4 | |
| 12 | L. orientalis | DQ107542 | 124 | 148 | 166 | 164 | 141 | 133 | 133 | 133 | 132 | 2 | 3 | 2.7 | 21.3 | 22.5 | |
| 13 | L. orientalis | DQ107544 | 125 | 149 | 167 | 165 | 142 | 135 | 135 | 135 | 134 | 20 | 21 | 22 | 20.7 | 22.0 | |
| 14 | L. chinensis | DQ107545 | 132 | 156 | 154 | 151 | 144 | 141 | 141 | 141 | 140 | 151 | 152 | 153 | 151 | 22.7 | |
| 15 | Lernaea cyprinacea | DQ107548 | 155 | 176 | 195 | 195 | 182 | 183 | 183 | 183 | 183 | 181 | 182 | 183 | 182 | 180 |
| Accession Number | OR232207 L. cleopatra | OR232208 L. clariae | OR232209 L. clariae | NC 025239 Lernaea cyprinacea | |
|---|---|---|---|---|---|
| L. cleopatra UL237 | OR232207 | 20.1 | 19.9 | 26.8 | |
| L. clariae UL241 | OR232208 | 137 | 0.1 | 26.2 | |
| L. clariae UL242 | OR232209 | 136 | 1 | 26.4 | |
| Lernaea cyprinacea | NC 025239 | 183 | 179 | 180 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rindoria, N.M.; Gichana, Z.; Morara, G.N.; van Wyk, C.; Smit, W.J.; Smit, N.J.; Luus-Powell, W.J. Scanning Electron Microscopy and First Molecular Data of Two Species of Lamproglena (Copepoda: Lernaeidae) from Labeo victorianus (Cyprinidae) and Clarias gariepinus (Clariidae) in Kenya. Pathogens 2023, 12, 980. https://doi.org/10.3390/pathogens12080980
Rindoria NM, Gichana Z, Morara GN, van Wyk C, Smit WJ, Smit NJ, Luus-Powell WJ. Scanning Electron Microscopy and First Molecular Data of Two Species of Lamproglena (Copepoda: Lernaeidae) from Labeo victorianus (Cyprinidae) and Clarias gariepinus (Clariidae) in Kenya. Pathogens. 2023; 12(8):980. https://doi.org/10.3390/pathogens12080980
Chicago/Turabian StyleRindoria, Nehemiah M., Zipporah Gichana, George N. Morara, Coret van Wyk, Willem J. Smit, Nico J. Smit, and Wilmien J. Luus-Powell. 2023. "Scanning Electron Microscopy and First Molecular Data of Two Species of Lamproglena (Copepoda: Lernaeidae) from Labeo victorianus (Cyprinidae) and Clarias gariepinus (Clariidae) in Kenya" Pathogens 12, no. 8: 980. https://doi.org/10.3390/pathogens12080980