Connectivity of Edaphic and Endolithic Microbial Niches in Cold Mountain Desert of Eastern Pamir (Tajikistan)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Field Area, Sampling and Study Design
2.2. Chemical Analysis of the Soil
2.3. Electron Microscopy Analysis with Energy Dispersive Spectroscopy (SEM-EDS) and Transmission Electron Microscopy (TEM)
2.4. Isolation, Cultivation and Morphological Analysis of Cyanobacteria
2.5. DNA Extraction, PCR and Illumina MiSeq Sequencing
2.6. Bioinformatic Analysis and Statistics
3. Results
3.1. Physicochemical Characteristics of Studied Mineral Substrates
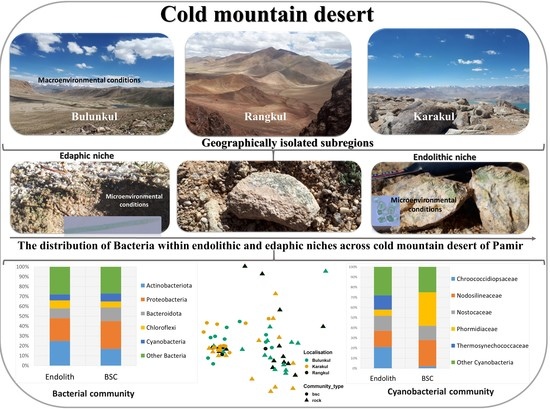
3.2. The Taxonomic Composition and Structure of Endolithic and Edaphic Communities
3.3. The Taxonomic Composition of Cyanobacterial Communities and Relative Abundance of Identified Taxa
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prăvălie, R. Drylands extent and environmental issues: A global approach. Earth Sci Rev. 2016, 161, 259–278. [Google Scholar] [CrossRef]
- Pointing, S.B.; Belnap, J. Microbial colonization and controls in dryland systems. Nat. Rev. Genet. 2012, 10, 551–562. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; D’Odorico, P.; Evans, J.P.; Eldridge, D.J.; McCabe, M.F.; Caylor, K.K.; King, E.G. Dryland ecohydrology and climate change: Critical issues and technical advances. Hydrol. Earth Syst. Sci. 2012, 16, 2585–2603. [Google Scholar] [CrossRef] [Green Version]
- Mętrak, M.; Sulwinski, M.; Chachulski, L.; Wilk, M.B.; Suska-Malawska, M. Creeping Environmental Problems in the Pamir Mountains: Landscape Conditions, Climate Change, Wise Use and Threats. In Climate Change Impacts on High—Altitude Ecosystems; Springer Science and Business Media LLC.: Berlin, Germany, 2015; pp. 665–694. [Google Scholar]
- Mętrak, M.; Chachulski, Ł.; Navruzshoev, D.; Pawlikowski, P.; Rojan, E.; Sulwiński, M.; Suska-Malawska, M. Nature’s Patchwork: How water sources and soil salinity determine the distribution and structure of halophytic plant communities in arid environments of the Eastern Pamir. PLoS ONE 2017, 12, e0174496. [Google Scholar] [CrossRef]
- Qu, E.B.; Omelon, C.R.; Oren, A.; Meslier, V.; Cowan, D.A.; Maggs-Kölling, G.; DiRuggiero, J. Trophic selective pressures organize the composition of endolithic microbial communities from global deserts. Front. Microbiol. 2019, 10, 2952. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.C.; Archer, S.D.J.; Boyle, R.H.; Lacap-Bugler, D.C.; Belnap, J.; Pointing, S.B. Niche filtering of bacteria in soil and rock habitats of the Colorado Plateau Desert, Utah, USA. Front. Microbiol. 2016, 7, 1489. [Google Scholar] [CrossRef]
- Wierzchos, J.; de los Ríos, A.; Ascaso, C. Microorganisms in desert rocks: The edge of life on Earth. Int. Microbiol. 2012, 15, 171–181. [Google Scholar]
- Golubic, I.F.S. The Lithobiontic Ecological Niche, with Special Reference to Microorganisms. J. Sediment. Res. 1981, 51, 475–478. [Google Scholar] [CrossRef]
- Khomutovska, N.; de los Ríos, A.; Jasser, I. Diversity and Colonization Strategies of Endolithic Cyanobacteria in the Cold Mountain Desert of Pamir. Microorganisms 2021, 9, 6. [Google Scholar] [CrossRef]
- Makhalanyane, T.P.; Valverde, A.; Gunnigle, E.; Frossard, A.; Ramond, J.-B.; Cowan, D.A. Microbial ecology of hot desert edaphic systems. FEMS Microbiol. Rev. 2015, 39, 203–221. [Google Scholar] [CrossRef]
- Choe, Y.-H.; Kim, M.; Lee, Y.K. Distinct Microbial Communities in Adjacent Rock and Soil Substrates on a High Arctic Polar Desert. Front. Microbiol. 2021, 11, 3392. [Google Scholar] [CrossRef]
- Knowles, E.J.; Castenholz, R.W. Effect of exogenous extracellular polysaccharides on the desiccation and freezing tolerance of rock-inhabiting phototrophic microorganisms. FEMS Microbiol. Ecol. 2008, 66, 261–270. [Google Scholar] [CrossRef] [Green Version]
- Wierzchos, J.; DiRuggiero, J.; Vítek, P.; Artieda, O.; Souza-Egipsy, V.; Škaloud, P.; Tisza, M.; Davila, A.F.; Vílchez, C.; Garbayo, I.; et al. Adaptation strategies of endolithic chlorophototrophs to survive the hyperarid and extreme solar radiation environment of the Atacama Desert. Front. Microbiol. 2015, 6, 934. [Google Scholar] [CrossRef] [Green Version]
- Meslier, V.; Casero, M.C.; Dailey, M.; Wierzchos, J.; Ascaso, C.; Artieda, O.; McCullough, P.R.; DiRuggiero, J. Fundamental drivers for endolithic microbial community assemblies in the hyperarid Atacama Desert. Environ. Microbiol. 2018, 20, 1765–1781. [Google Scholar] [CrossRef] [Green Version]
- Rego, A.; Raio, F.; Martins, T.P.; Ribeiro, H.; Sousa, A.G.G.; Séneca, J.; Baptista, M.S.; Lee, C.K.; Cary, S.C.; Ramos, V.; et al. Actinobacteria and Cyanobacteria Diversity in Terrestrial Antarctic Microenvironments Evaluated by Culture-Dependent and Independent Methods. Front. Microbiol. 2019, 10, 1018. [Google Scholar] [CrossRef]
- Garrido-Benavent, I.; Pérez-Ortega, S.; Durán, J.; Ascaso, C.; Pointing, S.B.; Rodríguez-Cielos, R.; Navarro, F.; de los Ríos, A. Differential colonization and succession of microbial communities in rock and soil substrates on a maritime Antarctic glacier forefield. Front. Microbiol. 2020, 11, 126. [Google Scholar] [CrossRef] [Green Version]
- Durán, J.; Rodríguez, A.; Heiðmarsson, S.; Lehmann, J.R.K.; del Moral, A.; Garrido-Benavent, I.; de los Ríos, A. Cryptogamic cover determines soil attributes and functioning in polar terrestrial ecosystems. Sci. Total Environ. 2021, 762, 143169. [Google Scholar] [CrossRef]
- Christmas, N.A.M.; Anesio, A.M.; Sánchez-Baracaldo, P. Multiple adaptations to polar and alpine environments within cyanobacteria: A phylogenomic and Bayesian approach. Front. Microbiol. 2015, 6, 1070. [Google Scholar] [CrossRef] [Green Version]
- Van Goethem, M.W.; Makhalanyane, T.P.; Valverde, A.; Cary, S.C.; Cowan, D.A. Characterization of bacterial communities in lithobionts and soil niches from Victoria Valley, Antarctica. FEMS Microbiol. Ecol. 2016, 92, fiw051. [Google Scholar] [CrossRef] [Green Version]
- Van Goethem, M.W.; Cowan, D.A. Role of Cyanobacteria in the Ecology of Polar Environments. In Springer Polar Sciences; Springer Science and Business Media LLC.: Berlin, Germany, 2019; pp. 3–23. [Google Scholar]
- Mergelov, N.; Mueller, C.W.; Prater, I.; Shorkunov, I.; Dolgikh, A.; Zazovskaya, E.; Shishkov, V.; Krupskaya, V.; Abrosimov, K.; Cherkinsky, A.; et al. Alteration of rocks by endolithic organisms is one of the pathways for the beginning of soils on Earth. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Kleinteich, J.; Puddick, J.; Wood, S.A.; Hildebrand, F.; Laughinghouse, H.D.; Pearce, D.A.; Dietrich, D.; Wilmotte, A. Toxic cyanobacteria in svalbard: Chemical diversity of microcystins detected using a liquid chromatography mass spectrometry precursor ion screening method. Toxins 2018, 10, 147. [Google Scholar] [CrossRef] [Green Version]
- Gkelis, S.; Panou, M.; Konstantinou, D.; Apostolidis, P.; Kasampali, A.; Papadimitriou, S.; Kati, D.; Di Lorenzo, G.M.; Ioakeim, S.; Zervou, S.-K.; et al. Diversity, Cyanotoxin Production, and Bioactivities of Cyanobacteria Isolated from Freshwaters of Greece. Toxins 2019, 11, 436. [Google Scholar] [CrossRef] [Green Version]
- Khomutovska, N.; Sandzewicz, M.; Łach, Ł.; Malawska, M.S.; Chmielewska, M.; Marzec, H.M.; Cegłowska, M.; Niyatbekov, T.; Wood, S.A.; Puddick, J.; et al. limited microcystin, anatoxin and cylindrospermopsin production by cyanobacteria from microbial mats in cold deserts. Toxins 2020, 12, 244. [Google Scholar] [CrossRef] [Green Version]
- Chan, Y.; Lacap, D.C.; Lau, M.C.; Ha, K.Y.; Warren-Rhodes, K.A.; Cockell, C.S.; Cowan, D.A.; McKay, C.P.; Pointing, S.B. Hypolithic microbial communities: Between a rock and a hard place. Environ. Microbiol. 2012, 14, 2272–2282. [Google Scholar] [CrossRef]
- Chamizo, S.; Mugnai, G.; Rossi, F.; Certini, G.; De Philippis, R. Cyanobacteria Inoculation Improves Soil Stability and Fertility on Different Textured Soils: Gaining Insights for Applicability in Soil Restoration. Front. Environ. Sci. 2018, 6, 49. [Google Scholar] [CrossRef]
- Khomutovska, N.; Jerzak, M.; Kostrzewska-Szlakowska, I.; Kwiatowski, J.; Suska-Malawska, M.; Syczewski, M.; Jasser, I. Life in extreme habitats: Diversity of endolithic microorganisms from cold desert ecosystems of Eastern Pamir. Pol. J. Ecol. 2017, 65, 303–319. [Google Scholar] [CrossRef]
- Vanselow, K.A. The High-Mountain Pastures of the Eastern Pamirs (Tajikistan)—An Evaluation of the Ecological Basis and the Pasture Potential. Available online: http://d-nb.info/1010705040/ (accessed on 20 April 2020).
- Kabala, C.; Chachulski, Ł.; Gądek, B.; Korabiewski, B.; Mętrak, M.; Suska-Malawska, M. Soil development and spatial differentiation in a glacial river valley under cold and extremely arid climate of East Pamir Mountains. Sci. Total Environ. 2021, 758, 144308. [Google Scholar] [CrossRef]
- Jasser, I.; Kostrzewska-Szlakowska, I.; Kwiatowski, J.; Navruzshoev, D.; Suska-Malawska, M.; Khomutovska, N. Morphological and molecular diversity of benthic cyanobacteria communities versus environmental conditions in shallow, high mountain water bodies in eastern Pamir mountains (Tajikistan). Pol. J. Ecol. 2020, 67, 286–304. [Google Scholar] [CrossRef]
- Coleine, C.; Stajich, J.E.; Pombubpa, N.; Zucconi, L.; Onofri, S.; Selbmann, L. Sampling strategies to assess microbial diversity of Antarctic cryptoendolithic communities. Polar Biol. 2020, 43, 225–235. [Google Scholar] [CrossRef]
- Sulwiński, M.; Mętrak, M.; Wilk, M.; Suska-Malawska, M. Smouldering fire in a nutrient-limited wetland ecosystem: Long-lasting changes in water and soil chemistry facilitate shrub expansion into a drained burned fen. Sci. Total Environ. 2020, 746, 141142. [Google Scholar] [CrossRef]
- Ostrowska, A.; Gawliński, S.; Szczubiałka, Z. Metody Analizy i Oceny Właściwości Gleb i Roślin; Instytut Ochrony Środowiska: Warsaw, Poland, 1991. (In Polish) [Google Scholar]
- Klapetek, P.; Ohlídal, I.; Franta, D.; Montaigne-Ramil, A.; Bonanni, A.; Stifter, D.; Sitter, H. Atomic force microscopy characterization of ZnTe epitaxial films. Acta Phys. Slovaca 2003, 53, 223–230. [Google Scholar] [CrossRef]
- de los Ríos, A.; Ascaso, C. Preparative Techniques for Transmission Electron Microscopy and Confocal Laser Scanning Microscopy of Lichens. In Protocols in Lichenology. Culturing, Biochemistry, Ecophysiology and Use in Biomonitoring; Kranner, I.C., Beckett, R.P., Varma, A.K., Eds.; Springer: Berlin/Heidelberg, Germany, 2002; pp. 87–117. [Google Scholar]
- Guillard, R.L.; Lorenzen, C.J. Yellow-green algae with chlorophyllide. J. Phycol. 1972, 8, 10–14. [Google Scholar] [CrossRef]
- Rippka, R.; Stanier, R.Y.; Deruelles, J.; Herdman, M.; Waterbury, J.B. Generic Assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology 1979, 111, 1–61. [Google Scholar] [CrossRef] [Green Version]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2012, 41, e1. [Google Scholar] [CrossRef]
- Nübel, U.F.; Garcia-Pichel, M.G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 1997, 63, 3327–3332. [Google Scholar] [CrossRef] [Green Version]
- Wilmotte, A.; Van der Rauwera, G.; De Wachter, R. Structure of the 16-S ribosomal RNA of the thermophilic cyanobacterium chlorogloeopsis HTF (Mastigocladus laminosus HTF’) strain PCC7518, and phylogenetic analysis. FEBS Lett. 1993, 317, 96–100. [Google Scholar] [CrossRef] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Bokulich, N.A.; Kaehler, B.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome 2018, 6, 1–17. [Google Scholar] [CrossRef]
- Roush, D.; Giraldo-Silva, A.; Fernandes, V.M.C.; Maria Machado de Lima, N.; McClintock, S.; Velasco Ayuso, S.; Klicki, K.; Dirks, B.; Arantes Gama, W.; Sorochkina, K.; et al. Cydrasil: A Comprehensive Phylogenetic Tree of Cyanobacterial 16s rRNA Gene Sequences. Available online: https://github.com/FGPLab/cydrasil (accessed on 3 March 2021).
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Atschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Kolbe, D.L.; Eddy, S.R. Infernal 1.0: Inference of RNA alignments. Bioinformatics 2009, 25, 1335–1337. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML Version 8: A tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria; Available online: http://www.R-project (accessed on 13 January 2021).
- Wong, F.K.Y.; Lau, M.; Lacap, D.C.; Aitchison, J.C.; Cowan, D.A.; Pointing, S.B. Endolithic Microbial Colonization of Limestone in a High-altitude Arid Environment. Microb. Ecol. 2009, 59, 689–699. [Google Scholar] [CrossRef]
- Cary, S.C.; McDonald, I.R.; Barrett, J.E.; Cowan, D. A3 on the rocks: The microbiology of Antarctic dry valley soils. Nat. Rev. Microbiol. 2010, 8, 129–138. [Google Scholar] [CrossRef]
- Crits-Christoph, A.; Robinson, C.K.; Ma, B.; Ravel, J.; Wierzchos, J.; Ascaso, C.; Artieda, O.; Souza-Egipsy, V.; Casero, M.C.; DiRuggiero, J. Phylogenetic and functional substrate specificity for endolithic microbial communities in hyper-arid environments. Front. Microbiol. 2016, 7, 301. [Google Scholar] [CrossRef] [Green Version]
- Jung, P.; Schermer, M.; Briegel-Williams, L.; Baumann, K.; Lein-weber, P.; Karsten, U.; Lehnert, L.; Achilles, S.; Bendix, J.; Budel, B. Water availability shapes edaphic and lithic cyanobacterial communities in the Atacama Desert. J. Phycol. 2019, 55, 1306–1318. [Google Scholar] [CrossRef] [Green Version]
- Strunecký, O.; Elster, J.; Komárek, J. Taxonomic revision of the freshwater cyanobacterium “Phormidium” murrayi = Wilmottia murrayi. Fottea 2011, 11, 57–71. [Google Scholar] [CrossRef] [Green Version]
- Boison, G.; Mergel, A.; Jolkver, H.; Bothe, H. Bacterial life and dinitrogen fixation at a gypsum rock. Appl Environ. Microbiol. 2004, 70, 7070–7077. [Google Scholar] [CrossRef] [Green Version]
- Perkerson, R., III; Johansen, J.; Kovacik, L.; Brand, J.; Kaštovský, J.; Casamatta, D. A unique Pseudanabaenalean (Cyanobacteria) genus Nodosilinea gen. nov. based on morphological and molecular data. J. Phycol. 2011, 47, 1397–1412. [Google Scholar] [CrossRef]
- Malešević, T.P.; Dulić, T.; Obreht, I.; Trivunović, Z.; Marković, R.; Kostić, B.; Važić, T.; Meriluoto, J.; Svircev, Z. Cyanobacterial Potential for Restoration of Loess Surfaces through Artificially Induced Biocrusts. Appl. Sci. 2021, 11, 66. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khomutovska, N.; de los Ríos, A.; Syczewski, M.D.; Jasser, I. Connectivity of Edaphic and Endolithic Microbial Niches in Cold Mountain Desert of Eastern Pamir (Tajikistan). Biology 2021, 10, 314. https://doi.org/10.3390/biology10040314
Khomutovska N, de los Ríos A, Syczewski MD, Jasser I. Connectivity of Edaphic and Endolithic Microbial Niches in Cold Mountain Desert of Eastern Pamir (Tajikistan). Biology. 2021; 10(4):314. https://doi.org/10.3390/biology10040314
Chicago/Turabian StyleKhomutovska, Nataliia, Asunción de los Ríos, Marcin D. Syczewski, and Iwona Jasser. 2021. "Connectivity of Edaphic and Endolithic Microbial Niches in Cold Mountain Desert of Eastern Pamir (Tajikistan)" Biology 10, no. 4: 314. https://doi.org/10.3390/biology10040314