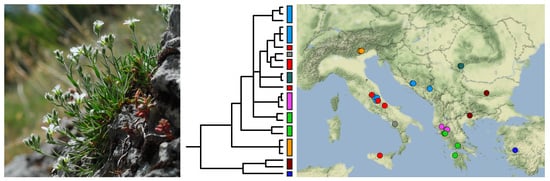
Relationships within Mcneillia Indicate a Complex Evolutionary History and Reveal a New Species of Minuartiella (Caryophyllaceae, Alsinoideae)
Abstract
:1. Introduction
2. Results
2.1. ITS Phylogeny
2.2. Characteristics of the Final Dataset
2.3. Nuclear and Chloroplast Phylogenies
2.4. Haplotype Network
3. Discussion
4. Taxonomic Treatment
- =
- Arenaria arduinoi var. italica Vis., Stirp. Dalmat. Spec.: 8. 1826 ≡ Minuartia graminifolia [“Rasse”] italica (Vis.) Graebn. in Ascherson & Graebner, Syn. Mitteleur. Fl. 5(1): 762. 1918 ≡ Minuartia graminifolia subsp. rosanoi var. italica (Vis.) Mattf. in Beibl. Bot. Jahrb. Syst. 126: 31. 1921. Type: Not designated.
- −
- “Alsine graminifolia var. hirsuta” Vis., Fl. Dalmat. 3: 178. 1852, nom. inval. (Art. 26.2 of ICN).
- −
- “Alsine graminifolia var. typica” Beck in Ann. Naturhist. Hofmus. 6: 324. 1891, nom. inval. (Art. 24.3 of ICN).
- =
- Arenaria arduinoi Vis., Stirp. Dalmat. Spec.: 8. 1826 (sub A. arduini, Art. 60.8 of ICN), nom. illeg. (Art. 52.2 of ICN) ≡ Alsine arduinoi (Vis.) Fenzl, Vers. Darstell. Alsin.: 57 (in tab.). 1833—Type (lectotype): [Republic of Croatia,] “E monti Biokovo [= from Mt. Biokovo] in Dalmatia”, IX s. a., [R.] Visiani s. n. (G!, individual in the middle), designated by Conti in Bot. J. Linn. Soc. 143: 426. 2003.
- =
- Arenaria arduinoi var. dalmatica Vis., Stirp. Dalmat. Spec.: 8. 1826 (sub A. Arduini var. dalmatica) ≡ Alsine rosanoi var. dalmatica (Vis.) Guss., Fl. Sicul. Syn. 1: 498. 1843 (“1842”) ≡ Alsine graminifolia var. dalmatica Beck in Ann. Naturhist. Hofmus. 6: 323. 1891—Type: Not designated.
- =
- Arenaria alpicola Ten., Fl. Napol. 4: 224. 1830—Type (lectotype): [Italy,] “Monte dei Fiori, Pizzo di Sivo, Majella | Costone nella discesa verso il Campiglione”, s. d., s. c., s. n. (NAP, barcode NAP000259-B!, sub Arenaria Rosani, A. alpicola), designated by Conti & Santangelo in Taxon 50: 193. 2001.
- =
- Alsine graminifolia var. glaberrima Vis., Fl. Dalmat. 3: 178. 1852 ≡ Minuartia graminifolia var. glaberrima (Vis.) Hayek in Denkschr. Akad. Wiss. Wien, Math.-Naturwiss. Kl. 94: 135. 1917—Type: Not designated.
- =
- Alsine graminifolia var. semiglabra Vis., Fl. Dalmat. 3: 178. 1852—Type: Not designated.
- =
- Alsine graminifolia var. dinarica Beck in Ann. Naturhist. Hofmus. 6: 324. 1891 ≡ Minuartia clandestina f. dinarica (Beck) Trinajstić, Suppl. Fl. Anal. Jugosl. 5: 6. 1978—Type: Not designated.
- =
- Alsine graminifolia var. dinarica f. subglabra Beck in Ann. Naturhist. Hofmus. 6: 324. 1891 ≡ Minuartia graminifolia [“Rasse”] clandestina var. dinarica f. subglabra (Beck) Graebn. in Ascherson & Graebner, Syn. Mitteleur. Fl. 5(1): 763. 1918 ≡ Minuartia clandestina f. subglabra (Beck) Trinajstić, Suppl. Fl. Anal. Jugosl. 5: 6. 1978—Type: Not designated.
- −
- −
- “Minuartia saxifraga subsp. rumelica” Mattf. in Bot. Jahrb. Syst. 57(2, Beibl. 126): 31, nom. inval. (Art. 26.2 of ICN).
- −
- “Minuartia saxifraga subsp. tmolea” Mattf. in Bot. Jahrb. Syst. 57(2, Beibl. 126): 31, nom. inval. (Art. 38.1 of ICN).
5. Materials and Methods
5.1. Sample Collection and DNA Extraction
5.2. Marker Selection, Amplification, and Sequencing
5.3. Sequence Alignment and Exploratory Data Analysis
5.4. Analysis of the Final Datasets
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fernanda Mazine, F.; Quintino Faria, J.E.; Giaretta, A.; Vasconcelos, T.; Forest, F.; Lucas, E. Phylogeny and biogeography of the hyper–diverse genus Eugenia (Myrtaceae: Myrteae), with emphasis on E. sect. Umbellatae, the most unmanageable clade. Taxon 2018, 67, 752–769. [Google Scholar] [CrossRef]
- Gargiulo, R.; Del Guacchio, E.; Caputo, P. Phylogenetic reconstruction of Asperula sect. Cynanchicae (Rubiaceae) reveals a mosaic of evolutionary histories. Taxon 2015, 64, 754–769. [Google Scholar] [CrossRef]
- Hilpold, A.; Vilatersana, R.; Susanna, A.; Meseguer, A.S.; Boršić, I.; Constantinidis, T.; Filigheddu, R.; Romaschenko, K.; Suárez-Santiago, V.N.; Tugay, O.; et al. Phylogeny of the Centaurea group (Centaurea, Compositae)—Geography is a better predictor than morphology. Mol. Phylogenet. Evol. 2014, 77, 195–215. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.C.; Nicolle, D.; Steane, D.A.; Vaillancourt, R.E.; Potts, B.M. High density, genome-wide markers and intra-specific replication yield an unprecedented phylogenetic reconstruction of a globally significant, speciose lineage of Eucalyptus. Mol. Phylogenet. Evol. 2016, 105, 63–85. [Google Scholar] [CrossRef]
- Santos, M.F.; Sano, P.T.; Forest, F.; Lucas, E. Phylogeny, morphology and circumscription of Myrcia sect. Sympodiomyrcia (Myrcia s.l., Myrtaceae). Taxon 2016, 65, 759–774. [Google Scholar] [CrossRef]
- Del Guacchio, E.; Caputo, P. Splitting Asperula (Rubiaceae): A proposal for consistency purposes within sections Cynanchicae, Thliphthisa and Hexaphylla. Plant Biosyst. 2020, 154, 766–782. [Google Scholar] [CrossRef]
- Mitka, J.; Novikov, A.; Rottensteiner, W.K. The taxonomic circumscription of Aconitum subgenus Aconitum (Ranunculaceae) in Europe. Webbia 2021, 76, 11–45. [Google Scholar] [CrossRef]
- Stone, R.D. The species-rich, paleotropical genus Memecylon (Melastomataceae): Molecular phylogenetics and revised infrageneric classification of the African species. Taxon 2014, 63, 539–561. [Google Scholar] [CrossRef]
- Pirani, A.; Zarre, S.; Pfeil, B.E.; Bertrand, Y.J.K.; Assadi, M.; Oxelman, B. Molecular phylogeny of Acanthophyllum (Caryophyllaceae: Caryophylleae), with emphasis on infrageneric classification. Taxon 2014, 63, 592–607. [Google Scholar] [CrossRef]
- Sadeghian, S.; Zarre, S.; Rabeler, R.K.; Heubl, G. Molecular phylogenetic analysis of Arenaria (Caryophyllaceae: Tribe Arenarieae) and its allies inferred from nuclear DNA internal transcribed spacer and plastid DNA rps16 sequences. Bot. J. Linn. Soc. 2015, 178, 648–669. [Google Scholar] [CrossRef]
- Frajman, B.; Heidari, N.; Oxelman, B. Phylogenetic relationships of Atocion and Viscaria (Sileneae, Caryophyllaceae) inferred from chloroplast, nuclear ribosomal, and low-copy gene DNA sequences. Taxon 2009, 58, 811–824. [Google Scholar] [CrossRef]
- Moore, A.J.; Messick, J.A.; Kadereit, J.W. Range and niche expansion through multiple interspecific hybridization: A genotyping by sequencing analysis of Cherleria (Caryophyllaceae). BMC Ecol. Evol. 2021, 21, 40. [Google Scholar] [CrossRef]
- Dillenberger, M.S.; Kadereit, J.W. Simultaneous speciation in the European high mountain flowering plant genus Facchinia (Minuartia s.l., Caryophyllaceae) revealed by genotyping-by-sequencing. Mol. Phylogenet. Evol. 2017, 112, 23–35. [Google Scholar] [CrossRef]
- Iamonico, D. Habrosia (Caryophyllaceae) a monotypic genus endemic to Western Asia: Morphological and molecular remarks. Acta Bot. Croat. 2021, 80, 208–214. [Google Scholar] [CrossRef]
- Frajman, B.; Eggens, F.; Oxelman, B. Hybrid origins and homoploid reticulate evolution within Heliosperma (Sileneae, Caryophyllaceae)—a multigene phylogenetic approach with relative dating. Syst. Biol. 2009, 58, 328–345. [Google Scholar] [CrossRef]
- Cires, E.; Prieto, J.A.F. Phylogenetic relationships of Petrocoptis A. Braun ex Endl. Caryophyllaceae), a discussed genus from the Iberian Peninsula. J. Plant Res. 2015, 128, 223–238. [Google Scholar] [CrossRef]
- Kool, A.; Bengtson, A.; Thulin, M. Polyphyly of Polycarpon (Caryophyllaceae) inferred from DNA sequence data. Taxon 2007, 56, 775–782. [Google Scholar] [CrossRef]
- Iamonico, D. Augustea (Polycarpaeae, Caryophyllaceae), a new genus from South America. Phytotaxa 2015, 236, 71–78. [Google Scholar] [CrossRef]
- Yao, G.; Xue, B.; Liu, K.; Li, Y.; Huang, J.; Zhai, J. Phylogenetic estimation and morphological evolution of Alsineae (Caryophyllaceae) shed new insight into the taxonomic status of the genus Pseudocerastium. Plant Divers. 2021, 43, 299–307. [Google Scholar] [CrossRef]
- Zhang, M.L.; Zeng, X.Q.; Li, C.; Sanderson, S.C.; Byalt, V.V.; Lei, Y. Molecular phylogenetic analysis and character evolution in Pseudostellaria (Caryophyllaceae) and description of a new genus, Hartmaniella, in North America. Bot. J. Linn. Soc. 2017, 184, 444–456. [Google Scholar] [CrossRef]
- Moiloa, N.A.; Mesbah, M.; Nylinder, S.; Manning, J.; Forest, F.; de Boer, H.J.; Bacon, C.D.; Oxelman, B. Biogeographic origins of southern African Silene (Caryophyllaceae). Mol. Phylogenet. Evol. 2021, 162, 107199. [Google Scholar] [CrossRef] [PubMed]
- Frajman, B.; Schönswetter, P.; Weiss-Schneeweiss, H.; Oxelman, B. Origin and diversification of South American polyploid Silene sect. Physolychnis (Caryophyllaceae) in the Andes and Patagonia. Front. Genet. 2018, 9, 639. [Google Scholar] [CrossRef] [PubMed]
- Rautenberg, A.; Hathaway, L.; Oxelman, B.; Prentice, H.C. Geographic and phylogenetic patterns in Silene section Melandrium (Caryophyllaceae) as inferred from chloroplast and nuclear DNA sequences. Mol. Phylogenet. Evol. 2010, 57, 978–991. [Google Scholar] [CrossRef] [PubMed]
- Sharples, M.T.; Tripp, E.A. RAD sequencing rejects a long-distance disjunction in Stellaria (Caryophyllaceae) and yields support for a new southern Rocky Mountains endemic. Taxon 2019, 68, 280–296. [Google Scholar] [CrossRef]
- Iamonico, D. Engellaria (Caryophyllaceae), a new North American genus segregated from Stellaria. Acta Bot. Mex. 2021, 128, e1846. [Google Scholar] [CrossRef]
- Dillenberger, M.S.; Kadereit, J.W. Maximum polyphyly: Multiple origins and delimitation with plesiomorphic characters require a new circumscription of Minuartia (Caryophyllaceae). Taxon 2014, 63, 64–88. [Google Scholar] [CrossRef]
- Mattfeld, J. Geographisch-genetische Untersuchungen über die Gattung Minuartia (L.) Hiern. Repert. Spec. Nov. Regni Veg. Beih. 1922, 15, 1–228. [Google Scholar]
- McNeill, J. Taxonomic studies in the Alsinoideae II: A revision of the species in the Orient. Notes Roy. Bot. Gard. Edinb. 1963, 24, 241–404. [Google Scholar]
- Halliday, G. Minuartia L. […] Sect. Lanceolatae (Fenzl) Graebner. In Flora Europaea, 2nd ed.; Tutin, T.G., Burges, N.A., Chater, A.O., Edmondson, J.R., Heywood, V.H., Moore, D.M., Valentine, D.H., Walters, S.M., Webb, D.A., Eds.; Cambridge University Press: Cambridge, UK, 1993; Volume 1, p. 157. [Google Scholar]
- Strid, A. Mountain Flora of Greece; Cambridge University Press: Cambridge, UK, 1986; Volume 1, pp. 92–107. [Google Scholar]
- POWO. Plants of the World Online. Facilitated by the Royal Botanic Gardens, Kew. Published on the Internet. Available online: http://www.plantsoftheworldonline.org/ (accessed on 25 May 2021).
- Bartolucci, F.; Conti, F.; Iamonico, D.; Del Guacchio, E. A new combination in Mcneillia (Caryophyllaceae) for the Italian flora. Phytotaxa 2014, 170, 139–140. [Google Scholar] [CrossRef]
- Conti, F. Minuartia graminifolia (Caryophyllaceae), a south-east European species. Bot. J. Linn. Soc. 2003, 143, 419–432. [Google Scholar] [CrossRef]
- Marhold, K. Caryophyllaceae. In: Euro + Med Plantbase—The Information Resource for Euro-Mediterranean Plant Diversity. 2011. Available online: https://europlusmed.org/cdm_dataportal/taxon/71ba4249-1991-4f98-afc7-4ef7e3b2a31b (accessed on 21 May 2021).
- Koç, M.; Hamzaoğlu, E.; Aksoy, A. Morphological and molecular evidence of Turkish Minuartiella species (Caryophyllaceae), with a description of a new species and a proposal for a new combination. Phytotaxa 2019, 409, 161–171. [Google Scholar] [CrossRef]
- Kamari, G. Minuartia. In Flora Hellenica; Strid, A., Tan, K., Eds.; Koeltz Scientific Books: Konigstein, Germany, 1997; Volume 1, pp. 170–191. [Google Scholar]
- McNeill, J. Minuartia. In Flora of Turkey and the East Aegean Islands; Davis, P.H., Ed.; Edinburgh University Press: Edinburgh, UK, 1967; Volume 2, pp. 38–67. [Google Scholar]
- Wolf, P.G.; Murray, R.A.; Sipes, S.D. Species-independent, geographical structuring of chloroplast DNA haplotypes in a montane herb Ipomopsis (Polemoniaceae). Mol. Ecol. 1997, 6, 283–291. [Google Scholar] [CrossRef]
- Okuyama, Y.; Fujii, N.; Wakabayashi, M.; Kawakita, A.; Ito, M.; Watanabe, M.; Murakami, N.; Makoto, K. Nonuniform concerted evolution and chloroplast capture: Heterogeneity of observed introgression patterns in three molecular data partition phylogenies of Asian Mitella (Saxifragaceae). Mol. Biol. Evol. 2005, 22, 285–296. [Google Scholar] [CrossRef]
- Du Pasquier, P.E.; Jeanmonod, D.; Naciri, Y. Morphological convergence in the recently diversified Silene gigantea complex (Caryophyllaceae) in the Balkan Peninsula and south-western Turkey, with the description of a new subspecies. Bot. J. Linn. Soc. 2017, 183, 474–493. [Google Scholar] [CrossRef]
- Alban, D.M.; Biersma, E.M.; Kadereit, J.W.; Dillenberger, M. Colonization of the Southern Hemisphere by Sagina and Colobanthus (Caryophyllaceae). Plant Syst. Evol. 2022, 308, 1. [Google Scholar] [CrossRef]
- Gussone, G. Florae Siculae Synopsis; Ex Typis Tramater: Neapoli, Italy, 1843; Volume 1, p. 923. [Google Scholar]
- De Halácsy, E. Conspectus Florae Graecae; Sumptibus Guilelmi Engelmann: Lipsiae, Germany, 1901. [Google Scholar]
- Turland, N.J.; Wiersema, J.H.; Barrie, F.R.; Greuter, W.; Hawksworth, D.L.; Herendeen, P.S.; Knapp, S.; Kusber, W.H.; Li, D.Z.; Marhold, K.; et al. International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017. In Regnum Vegetabile; Koeltz Botanical Books: Glashütten, Germany, 2018; Volume 159, pp. 1–254. [Google Scholar] [CrossRef]
- Bluff, M.J.; Nees, C.G.; Schauer, G.C. Compendium Florae Germanicae, 2nd ed.; Sumptibus, J.L., Ed.; Schrag: Norimbergae, Germany, 1837; Volume 1, p. 448. [Google Scholar]
- De Visiani, R. Flora Dalmatica; Apud Friedericum Hofmeister: Lipsiae, Germany, 1852; Volume 3, p. 390. [Google Scholar]
- Beck von Mannagetta, G.R. Flora von Südbosnien und der angrenzenden Hercegovina (VI Teil). Ann. Naturhist. Hofmus. 1891, 6, 307–344. [Google Scholar]
- Kováts, D. Plant types of Sándor Jávorka in the Hungarian Natural History Museum in Budapest III. Ann. Hist. Nat. Mus. Nat. Hung. 2000, 92, 21–40. [Google Scholar]
- Reich, D.; Gutermann, W.; Bardy, K.; Rainer, H.; Raus, T.; Sonnleitner, M.; Tan, K.; Lachmayer, M. The type specimens in Eugen von Halácsy’s Herbarium Graecum. Phytotaxa 2021, 493, 1–156. [Google Scholar] [CrossRef]
- Hiepko, P. The collections of the Botanical Museum Berlin-Dahlem (B) and their history. Englera 1987, 7, 219–252. [Google Scholar]
- De Visiani, R. Stirpium Dalmaticarum Specimen; Typis Crescinianis: Patavii, Italy, 1826; p. 57. [Google Scholar]
- Tenore, M. Flora Napolitana; Dalla Stamperia Francese: Napoli, Italy, 1830; Volume 2, p. 358. [Google Scholar]
- Mattfeld, J. Beitrag zur Kenntnis der systematischen Gliederung und geographischen Verbreitung der Gattung Minuartia. Bot. Jahrb. Syst. 1922, 57, 13–63. [Google Scholar]
- Stafleu, F.A.; Cowan, R.S. Taxonomic Literature, 2nd ed.; Scheltema and Holkema: Utrecht, The Netherlands, 1979; Volume 3, p. 991. [Google Scholar]
- Thiers, B. Index Herbariorum: A Global Directory of Public Herbaria and Associated Staff. New York Botanical Garden’s Virtual Herbarium. 2022. Available online: http://sweetgum.nybg.org/ih/ (accessed on 8 March 2021).
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. Version 3.51. 2018. Available online: http://www.mesquiteproject.org (accessed on 19 September 2021).
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nature Meth. 2012, 9, 772. [Google Scholar] [CrossRef]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Asmussen-Lange, C.B.; Hauser, T.P.; Deichmann, V.; Ørgaard, M. Hybridization and complex evolution of Barbarea vulgaris and related species (Brassicaceae). Mol. Phylogenet. Evol. 2022, 169, 107425. [Google Scholar] [CrossRef]
- Farris, J.S.; Källersjö, M.; Kluge, A.G.; Bult, C. Constructing a significance test for incongruence. Syst. Biol. 1995, 44, 570–572. [Google Scholar] [CrossRef]
- Farris, J.S.; Källersjö, M.; Kluge, A.G.; Bult, C. Testing significance of Incongruence. Cladistics 1995, 10, 315–319. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP* Phylogenetic Analysis using Parsimony (*and Other Methods), version 4; Sinauer Associates: Sunderland, MA, USA, 1998. [Google Scholar]
- Clement, M.; Snell, Q.; Walke, P.; Posada, D.; Crandall, K. TCS: Estimating gene genealogies. In Proceedings of the 16th International Parallel and Distributed Processing Symposium, Ft. Lauderdale, FL, USA, 15–19 April 2002; Volume 2, p. 184. [Google Scholar]
- Leigh, J.W.; Bryant, D. PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Templeton, A.R.; Crandall, K.A.; Sing, C.F. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 1992, 132, 619–633. [Google Scholar] [CrossRef]
- Linder, C.R.; Goertzen, L.R.; Heuvel, B.V.; Francisco-Ortega, J.; Jansen, R.K. The complete external transcribed spacer of 18S-26S rDNA: Amplification and phylogenetic utility at low taxonomic levels in Asteraceae and closely allied families. Mol. Phylogenet. Evol. 2000, 14, 285–303. [Google Scholar] [CrossRef]
- Moore, A.J.; Kadereit, J.W. The evolution of substrate differentiation in Minuartia series Laricifoliae (Caryophyllaceae) in the European Alps: In situ origin or repeated colonization? Am. J. Bot. 2013, 100, 2412–2425. [Google Scholar] [CrossRef]
- Aceto, S.; Caputo, P.; Cozzolino, S.; Gaudio, L.; Moretti, A. Phylogeny and evolution of Orchis and allied genera based on ITS DNA variation: Morphological gaps and molecular continuity. Mol. Phylogenet. Evol. 1999, 13, 67–76. [Google Scholar] [CrossRef]
- Ford, C.S.; Ayres, K.L.; Toomey, N.; Haider, N.; Van Alphen Stahl, J.; Kelly, L.J.; Wikström, N.; Hollingsworth, P.M.; Duff, R.J.; Hoot, S.B.; et al. Selection of candidate coding DNA barcoding regions for use on land plants. Bot. J. Linn. Soc. 2009, 159, 1–11. [Google Scholar] [CrossRef]
- Popp, M.; Oxelman, B. Inferring the history of the polyploid Silene aegaea (Caryophyllaceae) using plastid and homoeologous nuclear DNA sequences. Mol. Phylogenet. Evol. 2001, 20, 474–481. [Google Scholar] [CrossRef]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef]
- Sang, T.; Crawford, D.J.; Stuessy, T.F. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 1997, 84, 1120–1136. [Google Scholar] [CrossRef]
- Tate, J.A.; Simpson, B.B. Paraphyly of Tarasa (Malvaceae) and diverse origins of the polyploid species. Syst. Bot. 2003, 28, 723–737. [Google Scholar] [CrossRef]





| POWO [31] | Dillenberger and Kadereit [26] | Marhold [34] | Conti [33] | Halliday [29] |
|---|---|---|---|---|
| Mc. graminifolia subsp. brachypetala | Mc. graminifolia subsp. brachypetala | Mn. graminifolia subsp. brachypetala | Mn. graminifolia subsp. brachypetala | X |
| Mc. graminifolia subsp. clandestina | Mc. graminifolia subsp. clandestina | Mn. graminifolia subsp. clandestina | Mn. graminifolia subsp. clandestina | Mn. graminifolia subsp. clandestina |
| Mc. graminifolia subsp. graminifolia | Mc. graminifolia subsp. graminifolia | Mn. graminifolia subsp. graminifolia | Mn. graminifolia subsp. graminifolia | Mn. graminifolia subsp. graminifolia |
| Mc. graminifolia subsp. hungarica | - (1) | - (1) | Mn. graminifolia subsp. hungarica | - (1) |
| Mc. graminifolia subsp. rosanoi | - (1) | - (1) | Mn. graminifolia subsp. rosanoi | - (1) |
| Mc. moraldoi | Mc. moraldoi | X | Mn. moraldoi | X |
| Mc. pseudosaxifraga | Mc. pseudosaxifraga | Mn. pseudosaxifraga | Mn. pseudosaxifraga | Mn. pesudosaxifraga |
| Mc. saxifraga subsp. saxifraga | Mc. saxifraga subsp. saxifraga | Mn. saxifraga subsp. saxifraga | Mn. saxifraga subsp. saxifraga | Mn. saxifraga subsp. saxifraga |
| Mc. saxifraga subsp. tmolea | Mc. saxifraga subsp. tmolea | Mn. saxifraga subsp. tmolea | Mn. saxifraga subsp. tmolea | Mn. saxifraga subsp. tmolea |
| Mc. stellata | Mc. stellata | Mn. stellata | Mn. stellata | Mn. stellata |
| Taxon | N | Voucher Specimen |
|---|---|---|
| Mcneillia graminifolia subsp. brachypetala | 2 | GREECE. Mt. Boutsi * (APP nos. 36275 and 36277). |
| 1 | GREECE. Mt. Boutsi * (B, barcode B 10 0366304, isotype!). | |
| Mc. graminifolia subsp. clandestina | 1 | CROATIA. Mt. Biokovo * (FI, barcode FI066602). |
| 1 | CROATIA. Mt. Biokovo * (APP no. 58584). | |
| 1 | ITALY. Gran Sasso (APP no. 42385). | |
| 1 | ITALY. Montagna dei Fiori (APP no. 575). | |
| 1 | ITALY. Mt. Vettore (APP no. 62555). | |
| 1 | MONTENEGRO. Durmitor (B, barcode B 10 1092939). | |
| Mc. graminifolia subsp. graminifolia | 2 | ITALY. Vette di Feltre * (APP nos. 42396 and 42397). |
| 1 | ITALY. Vette di Feltre * (FI, barcode FI066605). | |
| Mc. graminifolia subsp. hungarica | 1 | ROMANIA. Mt. Arjana *, Fl. Romaniae Exsicc. 1221 (RO). |
| 1 | ROMANIA. Mt. Arjana *, Fl. Romaniae Exsicc. 1221b (RO). | |
| Mc. graminifolia subsp. rosanoi | 1 | ITALY. Gran Sasso (APP no. 42436). |
| 1 | ITALY. Mt. Secine (APP no. 42444). | |
| 1 | ITALY. Mt. Serrone (APP no. 42441). | |
| 1 | ITALY. Isnello (APP no. 42407). | |
| Mc. moraldoi | 1 | ITALY. Mt. Gelbison * (NAP, barcode NAP0000265). |
| Mc. pseudosaxifraga | 1 | ALBANIA. Mt. Nemercka (APP no. 48873). |
| 1 | GREECE. North Pindhos (APP nos. 36270). | |
| 1 | GREECE. Mt. Timphi (B, barcode B 10 1052716). | |
| Mc. saxifraga subsp. saxifraga | 1 | BULGARIA. Balkan * (B, barcode B 10 1092946). |
| 1 | GREECE. Mt. Kerkini, Greuter no. 16756 (C). | |
| Mc. saxifraga subsp. tmolea | 1 | TURKEY. Mt. Bozdağ * (FI, barcode FI066608). |
| Mc. stellata | 1 | GREECE. Mt. Erimanthos, Strid no. 33496 (C). |
| 1 | GREECE. Mt. Parnassos *, Baden & Franzen no. 940 (C). | |
| 2 | GREECE. Mt. Tsoumerka (APP nos. 43626 and 61429). | |
| Mn. recurva subsp. condensata | 1 | ITALY. Mt. Volturino (APP no. 63095). |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Luca, D.; Del Guacchio, E.; Conti, F.; Iamonico, D.; Caputo, P. Relationships within Mcneillia Indicate a Complex Evolutionary History and Reveal a New Species of Minuartiella (Caryophyllaceae, Alsinoideae). Plants 2022, 11, 2118. https://doi.org/10.3390/plants11162118
De Luca D, Del Guacchio E, Conti F, Iamonico D, Caputo P. Relationships within Mcneillia Indicate a Complex Evolutionary History and Reveal a New Species of Minuartiella (Caryophyllaceae, Alsinoideae). Plants. 2022; 11(16):2118. https://doi.org/10.3390/plants11162118
Chicago/Turabian StyleDe Luca, Daniele, Emanuele Del Guacchio, Fabio Conti, Duilio Iamonico, and Paolo Caputo. 2022. "Relationships within Mcneillia Indicate a Complex Evolutionary History and Reveal a New Species of Minuartiella (Caryophyllaceae, Alsinoideae)" Plants 11, no. 16: 2118. https://doi.org/10.3390/plants11162118