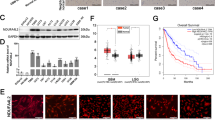
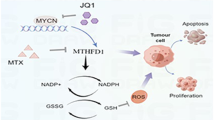
Abstract
Malate-aspartate shuttle (MAS) is essential for maintaining glycolysis and energy metabolism in tumors, while its regulatory mechanisms in neuroblastoma (NB), the commonest extracranial malignancy during childhood, still remain to be elucidated. Herein, by analyzing multi-omics data, GATA binding protein 2 (GATA2) and its antisense RNA 1 (GATA2-AS1) were identified to suppress MAS during NB progression. Mechanistic studies revealed that GATA2 inhibited the transcription of glutamic-oxaloacetic transaminase 2 (GOT2) and malate dehydrogenase 2 (MDH2). As a long non-coding RNA destabilized by RNA binding motif protein 15-mediated N6-methyladenosine methylation, GATA2-AS1 bound with far upstream element binding protein 3 (FUBP3) to repress its liquid-liquid phase separation and interaction with suppressor of zest 12 (SUZ12), resulting in decrease of SUZ12 activity and epigenetic up-regulation of GATA2 and other tumor suppressors. Rescue experiments revealed that GATA2-AS1 inhibited MAS and NB progression via repressing interaction between FUBP3 and SUZ12. Pre-clinically, administration of lentivirus carrying GATA2-AS1 suppressed MAS, aerobic glycolysis, and aggressive behaviors of NB xenografts. Notably, low GATA2-AS1 or GATA2 expression and high FUBP3, SUZ12, GOT2 or MDH2 levels were linked with unfavorable outcome of NB patients. These findings suggest that GATA2-AS1 inhibits FUBP3 phase separation to repress MAS and NB progression via modulating SUZ12 activity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
RNA-seq results have been deposited in GEO database (https://www.ncbi.nlm.nih.gov/geo, accession number GSE229665). Public datasets are available from GEO database (GSE62564, GSE45547), POSTAR3 (http://postar.ncrnalab.org), RBPDB (http://rbpdb.ccbr.utoronto.ca), ABLife (https://ablife.cc), BioGRID (https://thebiogrid.org), or PONDR (http://www.pondr.com) database.
References
Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–11.
Burns JS, Manda G. Metabolic pathways of the Warburg effect in health and disease: perspectives of choice, chain or chance. Int J Mol Sci. 2017;18:2755.
Gatenby RA, Gillies RJ. Why do cancers have high malate-aspartate shuttle? Nat Rev Cancer. 2004;4:891–9.
Borst P. The malate-aspartate shuttle (Borst cycle): How it started and developed into a major metabolic pathway. IUBMB Life. 2020;72:2241–59.
Pardo B, Contreras L, Satrústegui J. De novo synthesis of glial glutamate and glutamine in young mice requires aspartate provided by the neuronal mitochondrial aspartate-glutamate carrier Aralar/AGC1. Front Endocrinol. 2013;4:149.
Rub B, del Arco A, Bartley C, Satrustegui J, Maechler P. The malate-aspartate NADH shuttle member Aralar1 determines glucose metabolic fate, mitochondrial activity, and insulin secretion in beta cells. J Biol Chem. 2004;279:55659–66.
Wang C, Chen H, Zhang M, Zhang J, Wei X, Ying W. Malate-aspartate shuttle inhibitor aminooxyacetic acid leads to decreased intracellular ATP levels and altered cell cycle of C6 glioma cells by inhibiting glycolysis. Cancer Lett. 2016;378:1–7.
Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–61.
Ellis BC, Graham LD, Molloy PL. CRNDE, a long non-coding RNA responsive to insulin/IGF signaling, regulates genes involved in central metabolism. Biochim Biophys Acta. 2014;1843:372–86.
Rupaimoole R, Lee J, Haemmerle M, Ling H, Previs RA, Pradeep S, et al. Long noncoding RNA ceruloplasmin promotes cancer growth by altering glycolysis. Cell Rep. 2015;13:2395–402.
Kildisiute G, Kholosy WM, Young MD, Roberts K, Elmentaite R, van Hooff SR, et al. Tumor to normal single-cell mRNA comparisons reveal a pan-neuroblastoma cancer cell. Sci Adv. 2021;7:eabd3311.
Su Z, Fang H, Hong H, Shi L, Zhang W, Zhang W, et al. An investigation of biomarkers derived from legacy microarray data for their utility in the RNA-seq era. Genome Biol. 2014;15:523.
Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma’ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–44.
Kim GJ, Park SY, Kim H, Chun YH, Park SH. Chromosomal aberrations in neuroblastoma cell lines identified by cross species color banding and chromosome painting. Cancer Genet Cytogenet. 2001;129:10–16.
Gautier M, Thirant C, Delattre O, Janoueix-Lerosey I. Plasticity in neuroblastoma cell identity defines a noradrenergic-to-mesenchymal transition (NMT). Cancers. 2021;13:2904.
Yang H, Zhou L, Shi Q, Zhao Y, Lin H, Zhang M, et al. SIRT3-dependent GOT2 acetylation status affects the malate-aspartate NADH shuttle activity and pancreatic tumor growth. EMBO J. 2015;34:1110–25.
Han S, Zhu L, Zhu Y, Meng Y, Li J, Song P, et al. Targeting ATF4-dependent pro-survival autophagy to synergize glutaminolysis inhibition. Theranostics. 2021;11:8464–79.
Gameiro PA, Encheva V, Dos Santos MS, MacRae JI, Ule J. Metabolic turnover and dynamics of modified ribonucleosides by (13)C labeling. J Biol Chem. 2021;297:101294.
Agostini F, Zanzoni A, Klus P, Marchese D, Cirillo D, Tartaglia GG. catRAPID omics: a web server for large-scale prediction of protein-RNA interactions. Bioinformatics. 2013;29:2928–30.
Walia RR, Xue LC, Wilkins K, El-Manzalawy Y, Dobbs D, Honavar V. RNABindRPlus: a predictor that combines machine learning and sequence homology-based methods to improve the reliability of predicted RNA-binding residues in proteins. PLoS One. 2014;9:e97725.
Stark C, Breitkreutz B-J, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucl Acids Res. 2006;34:D535–39.
Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071.
Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67.
Ulianov SV, Velichko AK, Magnitov MD, Luzhin AV, Golov AK, Ovsyannikova N, et al. Suppression of liquid-liquid phase separation by 1,6-hexanediol partially compromises the 3D genome organization in living cells. Nucl Acids Res. 2021;49:10524–41.
Wang T, Kong S, Tao M, Ju S. The potential role of RNA N6-methyladenosine in cancer progression. Mol Cancer. 2020;19:88.
Zhao Z, Ju Q, Ji J, Li Y, Zhao Y. N6-Methyladenosine Methylation Regulator RBM15 is a Potential prognostic biomarker and promotes cell proliferation in pancreatic adenocarcinoma. Front Mol Biosci. 2022;9:842833.
Dong W, Bi J, Liu H, Yan D, He Q, Zhou Q, et al. Circular RNA ACVR2A suppresses bladder cancer cells proliferation and metastasis through miR-626/EYA4 axis. Mol Cancer. 2019;18:95.
Qutob N, Masuho I, Alon M, Emmanuel R, Cohen I, Di Pizio A, et al. RGS7 is recurrently mutated in melanoma and promotes migration and invasion of human cancer cells. Sci Rep. 2018;8:653.
Zhang L, Gao L, Shao M, Sun GYA. MYC target long non-coding RNA GATA2-AS1 regulates non-small cell lung cancer growth. Neoplasma. 2019;66:954–62.
Niu Y, Guo Y, Li Y, Shen S, Liang J, Guo W, et al. LncRNA GATA2-AS1 suppresses esophageal squamous cell carcinoma progression via the mir-940/PTPN12 axis. Exp Cell Res. 2022;416:113130.
Pan Y, Zhu Y, Zhang J, Jin L, Cao P. A feedback loop between GATA2-AS1 and GATA2 promotes colorectal cancer cell proliferation, invasion, epithelial-mesenchymal transition and stemness via recruiting DDX3X. J Transl Med. 2022;20:287.
Vicente C, Conchillo A, García-Sánchez MA, Odero MD. The role of the GATA2 transcription factor in normal and malignant hematopoiesis. Crit Rev Oncol Hematol. 2012;82:1–17.
Li YW, Wang JX, Yin X, Qiu SJ, Wu H, Liao R, et al. Decreased expression of GATA2 promoted proliferation, migration and invasion of HepG2 in vitro and correlated with poor prognosis of hepatocellular carcinoma. PLoS One. 2014;9:e87505.
Wang Y, He X, Ngeow J, Eng C. GATA2 negatively regulates PTEN by preventing nuclear translocation of androgen receptor and by androgen-independent suppression of PTEN transcription in breast cancer. Hum Mol Genet. 2012;21:569–76.
Böhm M, Locke WJ, Sutherland RL, Kench JG, Henshall SM. A role for GATA-2 in transition to an aggressive phenotype in prostate cancer through modulation of key androgen-regulated genes. Oncogene. 2009;28:3847–56.
Gau BH, Chen TM, Shih YHJ, Sun HS. FUBP3 interacts with FGF9 3’ microsatellite and positively regulates FGF9 translation. Nucl Acids Res. 2011;39:3582–93.
Huang HI, Chang YY, Lin JY, Kuo RL, Liu HP, Shih SR, et al. Interactome analysis of the EV71 5′ untranslated region in differentiated neuronal cells SH-SY5Y and regulatory role of FBP3 in viral replication. Proteomics. 2016;16:2351–62.
Weber A, Kristiansen I, Johannsen M, Oelrich B, Scholmann K, Gunia S, et al. The FUSE binding proteins FBP1 and FBP3 are potential c-myc regulators in renal, but not in prostate and bladder cancer. BMC Cancer. 2008;8:369.
Gao Q, Zhou R, Meng Y, Duan R, Wu L, Li R, et al. Long noncoding RNA CMPK2 promotes colorectal cancer progression by activating the FUBP3-c-Myc axis. Oncogene. 2020;39:3926–38.
Conway E, Healy E, Bracken AP. PRC2 mediated H3K27 methylations in cellular identity and cancer. Curr Opin Cell Biol. 2015;37:42–8.
Chammas P, Mocavini I, Di Croce L. Engaging chromatin: PRC2 structure meets function. Br J Cancer. 2020;122:315–28.
Mehta S, Zhang J. Liquid-liquid phase separation drives cellular function and dysfunction in cancer. Nat Rev Cancer. 2022;22:239–52.
Schaefer KN, Peifer M. Wnt/beta-catenin signaling regulation and a role for biomolecular condensates. Dev Cell. 2019;48:429–44.
Yamazaki T, Souquere S, Chujo T, Kobelke S, Chong YS, Fox AH, et al. Functional domains of NEAT1 architectural lncrna induce paraspeckle assembly through phase separation. Mol Cell. 2018;70:1038–53.
Wang R, Cao L, Thorne RF, Zhang XD, Li J, Shao F, et al. LncRNA GIRGL drives CAPRIN1-mediated phase separation to suppress glutaminase-1 translation under glutamine deprivation. Sci Adv. 2021;7:eabe5708.
Fang E, Wang X, Wang J, Hu A, Song H, Yang F, et al. Therapeutic targeting of YY1/MZF1 axis by MZF1-uPEP inhibits malate-aspartate shuttle and neuroblastoma progression. Theranostics. 2020;10:1555–71.
Zhao X, Li D, Yang F, Lian H, Wang J, Wang X, et al. Long noncoding RNA NHEG1 drives β-catenin transactivation and neuroblastoma progression through interacting with DDX5. Mol Ther. 2020;28:946–62.
Fang E, Wang X, Yang F, Hu A, Wang J, Li D, et al. Therapeutic targeting of MZF1-AS1/PARP1/E2F1 axis inhibits proline synthesis and neuroblastoma progression. Adv Sci. 2019;6:1900581.
Li D, Song H, Mei H, Fang E, Wang X, Yang F, et al. Armadillo repeat containing 12 promotes neuroblastoma progression through interaction with retinoblastoma binding protein 4. Nat Commun. 2018;9:2829.
Brauckhoff A, Malz M, Tschaharganeh D, Malek N, Weber A, Riener MO, et al. Nuclear expression of the ubiquitin ligase seven in absentia homolog (SIAH)-1 induces proliferation and migration of liver cancer cells. J Hepatol. 2011;55:1049–57.
Yang J, Smith DK, Ni H, Wu K, Huang D, Pan S, et al. SOX4-mediated repression of specific tRNAs inhibits proliferation of human glioblastoma cells. Proc Natl Acad Sci USA. 2020;117:5782–90.
Li H, Yang F, Hu A, Wang X, Fang E, Chen Y, et al. Therapeutic targeting of circ-CUX1/EWSR1/MAZ axis inhibits glycolysis and neuroblastoma progression. EMBO Mol Med. 2019;11:e10835.
Li D, Wang X, Mei H, Fang E, Ye L, Song H, et al. Long noncoding RNA pancEts-1 promotes neuroblastoma progression through hnRNPK-mediated β-catenin stabilization. Cancer Res. 2018;78:1169–83.
Son MJ, Ryu JS, Kim JY, Kwon Y, Chung KS, Mun SJ, et al. Upregulation of mitochondrial NAD(+) levels impairs the clonogenicity of SSEA1(+) glioblastoma tumor-initiating cells. Exp Mol Med. 2017;49:e344.
Acknowledgements
We are grateful for Dr. Kai Breuhahn for providing vectors. This work was granted by the National Natural Science Foundation of China (81903011, 82072801, 82173316, 82293663).
Author information
Authors and Affiliations
Contributions
XW and YG conceived and performed most of the experiments; GC, EF, JW, QL, DL, AH, BB, and YZ accomplished some of the in vitro experiments. XW, EF, HG, JS, and XD accomplished the in vivo studies. XW and EF undertook the mining of publicly available datasets. QT and LZ wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, X., Guo, Y., Chen, G. et al. Therapeutic targeting of FUBP3 phase separation by GATA2-AS1 inhibits malate-aspartate shuttle and neuroblastoma progression via modulating SUZ12 activity. Oncogene 42, 2673–2687 (2023). https://doi.org/10.1038/s41388-023-02798-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-023-02798-0