Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread to nearly every corner of the globe, causing societal instability. The resultant coronavirus disease 2019 (COVID-19) leads to fever, sore throat, cough, chest and muscle pain, dyspnoea, confusion, anosmia, ageusia and headache. These can progress to life-threatening respiratory insufficiency, also affecting the heart, kidney, liver and nervous systems. The diagnosis of SARS-CoV-2 infection is often confused with that of influenza and seasonal upper respiratory tract viral infections. Due to available treatment strategies and required containments, rapid diagnosis is mandated. This Review brings clarity to the rapidly growing body of available and in-development diagnostic tests, including nanomaterial-based tools. It serves as a resource guide for scientists, physicians, students and the public at large.
Similar content being viewed by others
Main
In the span of a few months, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was identified as the aetiological agent of coronavirus disease 2019 (COVID-19). Weeks later, viral diagnostic measures were deployed1. This served to supplement the common disease signs and symptoms of COVID-19 of cough, fever and dyspnoea. As all are seen during seasonal upper respiratory tract infections2, precise diagnostic tests detect viral nucleic acids, viral antigens or serological tests are required to affirm SARS-CoV-2 infection3. Chest computed tomography (CT) or magnetic resonance imaging (MRI) confirm disease manifestations2,3. The signature of COVID-19 is the life-threatening acute respiratory distress syndrome (ARDS)4. While the lung is the primary viral target, the cardiovascular, brain, kidney, liver and immune systems are commonly compromised by infection5. Thus, due to significant COVID-19 morbidity and mortality, containment of viral transmission through contact tracing, clinical assessment and virus detection was implemented through social distancing, face masks, contact isolation and hand hygiene to limit SARS-CoV-2 transmission6.
Overview of SARS-CoV-2 detection
The first step in managing COVID-19 is the rapid and accurate detection of SARS-CoV-2 enabled by real-time reverse transcription–polymerase chain reaction (RT–PCR)7. RT–PCR detects SARS-CoV-2 nucleic acids present in nasopharyngeal fluids8. Testing is used to prevent infectious spread between persons and communities that include asymptomatic infected persons, whose viral shedding can inadvertently spread the infection to the elderly and those with disease comorbidities9. Accurate viral detection is a starting point to contain the COVID-19 pandemic10. Lapses affect public safety, enabling infection spread aided by false-negative test results11. Improving test sensitivity and specificity remains an urgent need7. Serological testing complements virus detection, indicating past infection, which could be harnessed for therapeutic gain. Antibodies are detected by enzyme-linked immunosorbent assay using a qualitative detection of IgG or IgM antibodies12. Such tests determine an immune response against the viral spike (S) protein and may be helpful to assess protection against subsequent viral exposure and/or for contact tracing purposes13. Thus, the importance of such tests cannot be overstated. This is also true for epidemiological evaluations and broad global therapeutic needs14. Future work includes the development of diagnostic tests to improve immunoassay sensitivity and specificity13. Indeed, such testing will ultimately reveal viral protection as reinfections emerge15. Inducing immunity against SARS-CoV-2 is the next frontier for COVID-19 control15,16. To this end, our intent in this Review is to summarize the clinical disease presentation with a focus on how to best deploy nanomaterial-based and other diagnostic tests at individual, community and societal levels. The Review outlines current and future nanomaterial diagnostics for COVID-19. The intent is to facilitate the containment of the virus’s global spread12,15.
SARS-CoV-2 body fluid and tissue distribution
SARS-CoV-2 viral load and respiratory tract viral particles parallel virus dynamics in body fluids and tissue (Box 1). All affect concomitant host immune responses5,17. Viral load differs by sample, with respiratory, stool and serum samples showing broad variation in amounts of virus18. Spreading infection from the respiratory tract to other tissues and organs is linked to the cell-specific expression of angiotensin converting enzyme-2 (ACE2) receptors4. Viral load in respiratory samples is highest during the initial stages of the disease, reaching a peak in the second week, followed by lowered viral loads. In severe disease, the respiratory fluid virus is highest in the third and fourth weeks. In patients with comorbidities, viral persistence is continuous19, as highlighted from throat and anal swab sample assays20. Viral RT–PCR tests performed in throat swabs from disease-recovered individuals show positive results for up to 50 days and viral RNA was shown to be present in faecal and anal swabs weeks after respiratory samples were found negative20. Altogether, viral dynamics in hospitalized patients should be considered in recommendations for prevention and treatment of COVID-19.
Detection of SARS-CoV-2 viral shedding
In throat swabs and sputum, the viral shedding peaks at five to six days after symptom onset and ranges from 104 to 107 copies ml−1. This reflects higher virus levels in the respiratory tract21. The viral RNA detection rate in nasal swabs of infected people has approached 100%. The positivity rates for blood, saliva and tears are 88, 78 and 16%, respectively. The self-collection of naso- or oropharyngeal swabs facilitates large-scale population field testing employing the chemiluminescence immunoassay and the enzyme-linked immunosorbent and lateral-flow immunochromatographic assays22. The lateral-flow immunochromatographic assay uses gold nanoparticles (AuNPs) and a colorimetric label to provide a rapid platform for point-of-contact serological detection23. Here, SARS-CoV-2-specific antigen is conjugated with nanoparticles. By blood or saliva specimen loading, SARS-CoV-2 IgG and IgM can bind to the SARS-CoV-2 antigen and antibody, which is detected colorimetrically (Fig. 1). The assay is completed in 20 min with a ~90% accuracy24. To date, the minimum length of viral shedding is 7 d after symptom onset, with viral infectivity observed within 24 h (ref. 25). SARS-CoV-2 detection declines to undetectable levels, paralleling the presence of serum neutralizing antibodies25. Even among cases with concurrent high viral loads, the live virus could not be propagated in cell culture 8 d after symptom onset. These studies warrant the use of quantitative viral RNA load and serological assays when deciding whether to discontinue infection control precautions.
Commonly used immune-based tests contain SARS-CoV-2-specific recombinant antigens immobilized onto nitrocellulose membranes. Mouse anti-human IgM and IgG antibodies conjugated with coloured latex beads are immobilized on conjugate pads. The test sample contacts the membrane within the test. The coloured antibodies form latex conjugate complexes with human antiviral antibodies. This complex immobilized on the membrane is captured by the SARS-CoV-2-specific recombinant antigen. If SARS-CoV-2-specific IgG/IgM are present in the sample, this leads to a coloured band, indicating a positive test result. The complex is captured on the membrane by goat anti-mouse antibody, forming a red control line. A built-in control line appears in the test window. The absence of a coloured band demonstrates a negative result. a–e, The workflow begins with patient serum added to the sample flow well (S) (a), saline buffer is added dropwise (b), and the sample incubated (c) until antibody–antigen recognition (d) and SARS-CoV-2 antibody detection (e). f, The rabbit antibody–gold shows in the control (C) well. g, A positive test (T) band indicates the presence of COVID-19 antibody and results without a positive C band are invalid. Notably, this assay depicts a post-immune response and may show negative results for individuals who have been recently infected. It may also detect virus in previously infected but asymptomatic persons.
RT–PCR
Current diagnostic tests for the SARS-CoV-2 pandemic use nucleic acid, antibody and protein-based detections, but viral nucleic acid detection by RT–PCR remains the gold standard7. Nucleic acid tests have improved sensitivity and specificity for viral detection over the now available serological tests. The recognition of SARS-CoV-2 over common respiratory pathogens is contingent on RT–PCR serving as a sensitive, precise and specific viral detection. Despite the test’s accuracy, results have not yet enabled the containment of viral infection15. In February 2020, the US Food and Drug Administration (FDA) permitted licensed laboratories to report in-house SARS-CoV-2 diagnostic tests26. The procedure begins with the isolation and conversion of viral RNA to complementary DNA (cDNA). Next, the cDNA is amplified using Taq DNA polymerase. The RT–PCR test’s final overall workflow, which quantifies the viral load, is illustrated in Fig. 2a. The total turnaround time can exceed 2 d and runs the risk of reduced specificity through cross-contamination6. The tests are commonly performed in hospital laboratories7.
a, The RT–PCR assay. (i) A nasopharyngeal swab collects patient samples. (ii), (iii) RNA is extracted from fluids that contain SARS-CoV-2-infected cells and free viral particles. (iv) The recovered viral RNA is then reverse transcribed to cDNA and amplified for detection of viral nucleic acids. Conserved regions of the RdRp and E genes are the subgenomic viral segments amplified with a fluorogenic probe by qPCR. (v) Positive cases exceed the threshold of detection. b, The SARS-CoV-2 RT–LAMP assay: (i) amplification mixtures; (ii) RT–LAMP reaction; (iii) the products of SARS-CoV-2 RT–LAMP reactions. Although RT–PCR methods are used as the standard for detection of SARS-CoV-2 due to high sensitivity, limitations are present. As an alternative, isothermal amplification or LAMP was developed. When optimized for detection, the assay is as sensitive as standard PCR, detecting <10 viral copies per reaction. dNTP, deoxyribonucleoside 5′-triphosphate. c, SARS-CoV-2 saliva test. The illustration demonstrates SARS-CoV-2 infection in salivary glands and the released specific biomarkers that accumulate in the oral cavity. These are collected through a sample tube, tagged with a specific biomarker protein and run through lateral-flow rapid tests. Bulbar conjunctival injection in SARS-CoV-2 infected individuals is common.
Results from real-time RT–PCR using primers targeting different viral genome parts can be affected by viral RNA sequence variation. In addition, false-negative results may occur because of viral evolution16. Other limitations of RT–PCR tests include sample storage, low-quality nucleic acid purification, cost and wait times27. Despite such limitations, the RT–PCR test remains the gold standard for SARS-CoV-2 diagnostics. For the alternative in situ hybridization and immunohistochemistry, collection of large amounts of sample is required and this can generate aerosols and safety limitations28. Immunohistochemistry is dependent on the choice and specificity of the antibody and sample quality. The most definitive method for the virus is high-throughput sequencing, but this approach is limited due to the cost, equipment and skillsets required28.
Isothermal amplification is a useful alternative to thermal-cycling-based nucleic acid amplification29. Simplified RT–PCR is now available to detect diverse regions of the SARS-CoV-2 genome30. These detect the RNA-dependent S and RNA polymerase (RdRp)/helicase (Hel) proteins and the nucleocapsid (N) genes of SARS-CoV-2 (ref. 7). The RdRp/Hel assays are highly sensitive means for viral detection. This combined with proper handling of large sample numbers by automated solutions31 and cobas 6800 systems provides fast and reliable test results32.
RT loop-mediated isothermal amplification (RT–LAMP)
RT–LAMP is based on nanotechnology. LAMP-based diagnostic tests are detected by levels of turbidity or by colorimetric or fluorescence measures. This technique is simple to perform and visualize and has low background interference. The main limitations for LAMP testing involve experience, interpretation and reaction optimization33. Of two fluorescent dyes tested, the signal read-out properties of EvaGreen were superior to those of SYBR Green34. RT–LAMP is based on paper/strips integrated as part of a microfluidic platform to provide a lab-on-a-chip viral diagnosis35. In the test, fluorescein is assigned to one primer set and the product of the reaction catalysed by labelled RT29. An alternative method for LAMP accurately detects SARS-CoV-2 using a leucocrystal violet dye to provide a visible violet colour enabling detection of 100 copies per reaction. A means of improving the limit of detection of the LAMP assay is through a closed-tube Penn-RAMP, which combines RT–recombinant polymerase amplification and RT–LAMP in a single tube36. Figure 2b describes the RT–LAMP assay workflow. The products from three steps in the RT–LAMP system can serve as the template for the reaction of the LAMP system. In step (i) of Fig. 2b, solutions of deoxyribose adenosine triphosphate (dATP), polymerase (Bst 2.0) and avian myeloblastosis virus (AMV) transcriptase are used as LAMP reagents for preparing the amplification mixtures. The LAMP reagents’ reaction with biotin-labelled nucleoprotein (np)-backward loop primer (LB) (np-LB*) and fluorescein isothiocyanate (FITC)-labelled open reading frame 1a/b (F1ab)-forward loop primer (LF) (F1ab-LF*) starts the isothermal amplification (RT–LAMP reaction in step (ii)). Detectable COVID-19 RT–LAMP products are provided in step (iii). FITC/biotin-labelled np-LAMP and FITC/biotin-labelled F1ab-LAMP amplicons, the results of labelling F1ab-LF* and F1ab-LB* or np-LF* and np-LB* for digoxigenin and biotin, respectively29, are shown in step (iii). In contrast, FITC is assigned to the F1ab primer set; the F1ab-RT–LAMP product is labelled with FITC and biotin, while the np-RT–LAMP is labelled with digoxigenin and biotin.
Furthermore, the labelled F1ab-LF* and F1ab-LB* primers react under the optimized conditions, and SARS-CoV-2 RNA is converted to cDNA with AMV-RT at 63 °C in 40 min. This reaction provides the material for subsequent LAMP amplification; the RT–LAMP system consists of the FITC products and digoxigenin, for the detection of F1ab and np primer37. RNA extraction is time consuming and expensive, and requires centrifugation steps, which EasyCOV RT–LAMP tests do not require. EasyCOV technology is a simple and straightforward test without RNA extraction from the sample. The results of EasyCOV have demonstrated a sensitivity of 72.7%. LAMP techniques on saliva can identify people’s infection profiles. EasyCOV can detect SARS-CoV-2 in saliva and the test is viable for large-scale screenings of the general population due to its simple, fast and painless procedure for patients.
SARS-CoV-2 diagnostics using nanomaterials
Nanomaterial-based technology provides feasible alternatives to RT–PCR for quick and precise viral detection. For example, magnetic nanoparticles can facilitate viral RNA extraction through coprecipitation, followed by polyamine ester functionalization via (3-aminopropyl) triethoxysilane, and can be used for up to 50,000 diagnostic tests38. Quantum dots (QDs) could serve as ideal detection tools to study S protein–ACE2 binding dynamics and internalization due to their relatively small size, photostability and the ease of surface functionalization with biological molecules for Förster resonance energy transfer biosensing systems with various energy transfer partners39, such as AuNPs that are characterized by absorption of electromagnetic radiation in the visible region of the spectrum40. A colorimetric assay was developed based on thiol-modified antisense oligonucleotides conjugated with AuNPs for detection of SARS-CoV-2 N-gene RNA. This is used for rapid diagnosis and can be performed within 10 min. The lower limit of detection is 0.18 ng μl−1 RNA particles41. A recombinant S receptor binding domain conjugated to fluorescent QDs was created as an imaging probe for energy transfer quenching with ACE2-conjugated AuNPs. Upon binding of the S to the ACE2 receptor, fluorescence is quenched by the nearby AuNPs to enable monitoring of the binding events in the solution. QD probes can also facilitate cell-based assay identification and validation of inhibitors of the SARS-CoV-2 S protein and ACE2 receptor binding42. The QDs are used as probes to investigate other viral receptors43. This system can identify neutralizing antibodies and recombinant proteins for SARS-CoV-2 and other viruses with S-mediated cell recognition and entry.
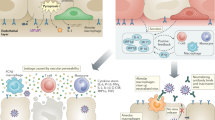
Biosensors have been developed for detecting influenza, the human immunodeficiency virus and other viral diseases44. Initially marred by low sensitivity and specificity, limitations were overcome by plasmonic (gold and silver), metal oxide nanoparticle and field effect transistor (FET) bio- and graphene sensors44,45. Graphene has wide application; it consists of hexagonal carbon structures arranged in a two-dimensional sheet. This gives it a large surface area, high electronic conductivity and high carrier mobility, and graphene biosensors are highly sensitive. When developing a graphene-based biosensor to detect SARS-CoV-2, coronavirus S antibody was immobilized on a graphene surface using 1-pyrenebutyric acid N-hydroxysuccinimide ester linkers. This graphene was used as a sensing material in a FET device to detect the S up to 1 fg ml−1 concentration46 (Fig. 3, top). The optical property of AuNPs and silver nanoparticles conjugated to antibodies, when they are bound to the viral antigens or RNA, causes a detectable signal, which can be used to detect SARS-CoV-2 (ref. 47). Toroidal plasmonic metasensors were developed that detect a femtomolar concentration of the viral S protein. They showed that monoclonal antibody conjugation on functionalized AuNPs could be detected up to 4.2 fM concentration (lower limit of detection). Transmission spectra of metasensors can shift the excitation with a polarized beam of light at terahertz frequency. Metasensors can be very useful in point of care (PoC) testing, where a rapid and sensitive assay is required48. Recently, researchers have devised a single-step, optical S-protein-specific nanoplasmonic resonance sensor that requires minimal sample preparation and provides fast and direct virus detection. In such a system, highly specific antibodies to SARS-CoV-2 were immobilized on nanosensor chip surfaces to which intact coronavirus particles bind through S protein, leading to plasmon resonance or intensity changes that can be optically measured through a sensing system49,50. For this assay, the lower limit of detection is 30 virus particles. The assay can be completed in 15 min. The assay can quantify virus below standard nasopharyngeal swab and saliva viral concentrations51. On analysing the specificity of the sensor for binding SARS-CoV-2 in comparison to SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV) and vesicular stomatitis pseudoviruses, nanoplasmonic sensor chips demonstrated very high specificity (>1,000:1) in detecting the SARS-CoV-2 (ref. 49). The nanoplasmonic sensor chips have the advantage of being low cost and scalable while maintaining uniformity and repeatability. The design of periodic nanostructures, without any external coupling optics52, allows sensor chips to be integrated with a standard 96-microwell plate or microfluidic cuvettes. This allows standard microplate reader measurements53. A low-cost, portable device controlled using a smartphone application can analyse SARS-CoV-2 in one step within 15 min with sensitive viral detection. Although the detection limit is 370, the virus can be quantified linearly from 0 to 107 virus particles per millilitre and it may find application in clinics, roadside screening sites and homes49. AuNP-based sensors coupled with artificial intelligence can detect volatile organic compounds associated with SARS-CoV-2 in exhaled breaths. The assay is able to detect virus on the basis of the change in resistance of the nanomaterial biosensor layer. The methods can be optimized in future months by using other nanomaterials and larger cohort testing54.
Representative illustration of nanomaterial-based biosensors for SARS-CoV-2 detection. Top: a FET-based biosensing device for detecting SARS-CoV-2. The sensor was produced using graphene sheets with specific antibodies against the SARS-CoV-2 S protein. Bottom: the dual-functional plasmonic photothermal biosensor and localized surface plasmon resonance biosensor on two-dimensional gold nanoislands functionalized with either cDNA receptors for detection of the selected SARS-CoV-2 sequences by fluorescence and FRET-based nucleic acid hybridization or with nucleic acid or antibody-functionalized nanomaterials for SARS-CoV-2 detection by colorimetric and antigen-binding assays. Ab, antibody; Ag, antigen; FRET, Förster resonance energy transfer; NPs, nanoparticles. Schematic ideas and technical methodological details were followed as represented in previously published reports36,37,45.
A clinical diagnostic sensor was developed that combines a dual-functional plasmonic photothermal effect with localized surface plasmon resonance sensing transduction. Tests are done on two-dimensional gold nanoislands (Fig. 3, bottom). The gold nanoislands contain complementary DNA receptors, which hybridize to SARS-CoV-2 nucleic acids. This system can be excited at two different wavelengths as it uses two different angles of incidence, one from a plasmonic photothermal biosensor and the other from localized surface plasmon resonance. It can detect RdRp-COVID, F1ab-COVID and envelope (E) genes from SARS-CoV-2. The dual-functional localized surface plasmon resonance biosensor has a lower detection limit of 0.22 pM and allows precise detection of selected SARS-CoV-2 sequences in a multigene mixture. The plasmonic sensing system can significantly reduce the rate of false-positive results55. Similarly, others developed a plasmonic nanohole array used to transmit light for the label-free detection of the pathogen in biological media without sample preparation. It can quantitate intact virions by capturing them on group-specific antiviral immunoglobulins immobilized at the surface of the sensor. The intact virus binds to a suspended nanohole array grating that couples incident light to surface plasmons, causing a redshift in surface plasmonic resonance frequency. The assay could detect small (vesicular stomatitis virus and pseudotyped Ebola) and large (vaccinia virus) enveloped viruses. The non-destructive nature of the assay allows for further analysis of progeny virions50. Overall, the biosensors and other material-science-based detection techniques can enable rapid and portable diagnostic SARS-CoV-2 testing.
Detection of SARS-CoV-2 antibodies
The synthesis of antibodies against SARS-CoV-2 is a primary immune response to infection. Neutralizing antibodies are found in up to 50% of infected individuals by day 7 and in all infected individuals by day 14. Serological studies are an alternative to RT–PCR for SARS-CoV-2 diagnostics. Combining real-time PCR and serological testing significantly increases positive viral detection rates. IgM levels increase during the first week after SARS‐CoV‐2 infection, peak after 2 weeks and then fall back to near-background levels in most individuals. IgG is detectable after 1 week and is maintained at a high level for a long period56. In contrast, IgG becomes detectable after 1 week, remains elevated for an extended period, sometimes even more than 48 d, and may serve to protect against reinfection. IgA responses appear between 4 and 10 d after infection. Notably, a diagnostic predictor is the presence of serum IgA57 as well as IgG and IgM58. The spectrum of SARS-CoV-2 antibodies is explained, in part, by divergent target antigens. Antibody titres can decrease 7 d after infection56. Recent studies have identified SARS-CoV-2-specific antibodies in the saliva59,60. Multiplex SARS-CoV-2 antibody immunoassays were investigated to determine differences between antibody levels in saliva and sera. Antibodies in saliva consistent with those in sera suggest parallel compartmental humoral immune responses60. A parallel study developed rapid immunoassay using the BreviTest platform technology for measuring salivary IgA, which correlates with COVID-19 disease severity.
Interestingly, low levels of IgA were seen in individuals with IgG without known exposure to the virus, and suggest that it may represent an indicator of herd immunity59. SARS-CoV-2-specific antibody detection, especially that in saliva, may be useful for surveillance. Questions remain as to which antigens are the best candidates for serological testing. While the viral S is perhaps the strongest candidate, what remains unresolved is what part of the S should be developed. Alternatively, multiple isoforms of the S protein, such as those found in variant strains, may be used to ensure assay reproducibility61. Time to results can vary from 13 min (Abbott ID NOW) to 45 min (Cepheid Xpert Xpress)62. Of the five antibody-based tests available, two are lateral-flow immunoassays (BioMedomics rapid test and SureScreen rapid test cassette), one is a time-resolved fluorescence immunoassay (Goldsite diagnostics kit) and two are colloidal gold immunoassays (Assay Genie rapid PoC kit and VivaDiag COVID-19 IgG–IgM based).
Clinical studies will be needed to determine their clinical relevance63. For N-based immunoassays, SARS-CoV-2 IgG (Abbott) shows a sensitivity of up to 100% (ref. 64). For S-based immunoassays, Liaison SARS-CoV-2 S1/S2 IgG and the combination S- and N-based platform COVID-19 VIRCLIA IgG MONOTEST demonstrated equivalent sensitivities. The plaque reduction neutralization test showed a sensitivity of 93.3%. To evaluate specificity, all of the tests except one, the enzyme-linked immunosorbent assay (IgG) (EUROIMMUN), produced at least one positive result for the negative SARS-CoV-2 antigen control. This probably represents large discrepancies between the testing platforms and the assay sensitivity relative to time. Although the plaque reduction neutralization test is the gold standard for immunoglobin-based detection, the test has constraints, including a limited number of sample analyses, and requires a biosafety level 3 laboratory. The titres obtained from the assays correlate well with the plaque reduction neutralization test. Currently, antibody assays are applied principally for epidemiological testing65.
SARS-CoV-2 antigens
A rapid diagnostic assay was also developed to detect the presence of viral antigens expressed by SARS-CoV-2 in samples from the respiratory tract of infected individuals66. For this assay, antigen present in the sample binds to antibodies affixed to a paper strip enclosed in a plastic casing. This reaction generates a visually detectable signal within half an hour. The detected antigen(s) are expressed only if the virus is actively replicating; therefore, the tests can be used to identify acute or early infection66. Also, a more common type of rapid diagnostic assay, which detects the presence of antibodies in the blood of infected individuals, has been marketed for COVID-19 by Abbott. Abbott’s test can detect the SARS-CoV-2 antibody on ARCHITECT i1000SR and i2000SR laboratory instruments, which can run ~100–200 tests per hour67. Antibodies against SARS-CoV-2 are produced after one week of infection68. The strength of any antibody response depends on age, nutritional status, disease severity, comorbid conditions and medications.
Saliva testing
The presence of SARS-CoV-2 RNA in saliva samples is not always associated with disease severity, compared to nasopharyngeal swabs. Nevertheless, human saliva has gained attention as an alternative diagnostic medium for detecting infections69. Naso- or oropharyngeal swabs show limitations in sample collection and present a risk to healthcare workers through sneeze or cough and transmission of virus particles by aerosols10. In addition, in cases of thrombocytopenia or any other coagulation disorders the collection procedure can precipitate bleeding. These complications have led to testing sputum collection for diagnostic purposes. Sputum is an easy directed and non-invasive method of sampling. However, one limitation is that 72% of individuals with COVID-19 are unable to produce sufficient sample volume18. As a multiconstituent oral fluid, saliva has demonstrated high potential for the surveillance of general health and disease70. The ease of collection for diagnostics and monitoring without the need for medical staff can lead to ease of sample collection (Fig. 2c). Saliva is a useful biological medium, as it comprises proteins, nucleic acids, electrolytes and hormones originating from multiple local and systemic sources. Saliva is known to contain approximately 30% of biomolecules found in blood and harbours viral microorganisms71. Moreover, saliva samples can be stored in stabilizing solutions and posted several days later in the testing centre. Saliva collection is less invasive to the donor than blood collection and can permit home sampling69,70. Analysis of saliva samples in individuals with COVID-19 may facilitate the detection of both the virus itself and the antibodies, and as such shows potential as a diagnostic medium. Human saliva sampling may have a major potential for COVID-19 screening51,72. There is a concordance between detecting respiratory pathogens, including two seasonal human coronaviruses, in saliva using RT–PCR22. Indeed, mean SARS-CoV-2 titres (virus copies ml−1) were five times higher (P < 0.05) in saliva (n = 37) compared with nasopharyngeal swabs (n = 46). Furthermore, none of the negative saliva samples became positive. In contrast, in five instances, nasopharyngeal swabs first tested negative for SARS-CoV-2, followed by a positive test result when repeated51,72. However, ever more reliable sample collection that can be self-administered is still needed, with a significant directive of current research activities.
Faecal tests
Knowledge regarding virus incubation, transmission and shedding is crucial for protecting healthcare professionals and stopping the spread of SARS-CoV-2. High incidence and viral persistence in faeces have been observed when nasopharyngeal swab samples were virus negative17. Notably, viral load in stool samples can be detected up to four weeks after disease onset. The risk of exposure of healthcare professionals to faecal material from infected individuals is well known, especially in highly aerosol-generative procedures. Facilities such as nursing homes may be particularly vulnerable to this pathway of infection transmission. While a high incidence of cough and fever are well established18, documented gastrointestinal symptoms support faecal–oral transmission routes73. On the basis of the prolonged viral shedding in faeces and respiratory samples 14 d after discharge, the European Centre for Disease Prevention has advocated continued self-isolation74. Studies have also shown that the live virus can be isolated from stool specimens17, supporting the possibility of faecal–oral transmission. As a result, evidence-based recommendations for gastrointestinal endoscopy and surgery are required where there may be an exposure risk to virus shedding in faeces. Finally, SARS-CoV-2 may be tracked through wastewater, which enables community surveillance and could be a powerful tool in tracking COVID-19 spread. There are now sewage screening tests for dormitories in an attempt to detect asymptomatic individuals. If positive, institutions can quarantine those infected to prevent subsequent SARS-CoV-2 outbreaks75.
Radiographic testing
Although quantitative and qualitative tests of viral nucleic acid RT–PCR tests are the primary assay for SARS-CoV-2 detection7,30, the sensitivities of these tests remain low for oropharyngeal (32%) and nasal (63%) swab samples21. RT–PCR tests can often take up to more than a week due to a shortage in testing supplies or lack of technical skills. Therefore, suspected cases, either with or without RT–PCR results, require additional affirmations. Combinations of radiographic, molecular and antigen-based assays have been used alone or in combination to determine the optimal means to make a definitive diagnosis of SARS-CoV-2 infection76. After the respiratory symptom presentation and nucleic acid viral detections, an initial evaluation of patients with COVID-19 commonly includes radiological examinations. Such examinations include a chest X-ray (CXR), CT or lung ultrasound (LUS). These, alone or together, can be also be used to stage SARS-CoV-2 infection76,77. Often, a simple CXR is sufficient. However, a negative CXR alone cannot rule out lung involvement78,79. While RT–PCR remains the gold standard for a virologic diagnosis, a CXR affords 69% sensitivity76,78,79. However, imaging features contained in a standard CXR are often non-specific. When radiographic features of the disease are seen they reflect dense radiological patches on the left upper lobe and lower corners of the lung. With disease progression, more well defined radiographic features are present and increase the veracity of a definitive COVID-19 diagnosis. However, while a CXR is the most useful test to affirm lung disease, it does not rule out alternative infections, especially in the context of presenting COVID-19 signs and symptoms, since it is not specific. Abnormalities such as pneumothorax, pulmonary oedema, pleural effusions, lung mass or lung collapse are alternatives79. The value of the CXR is further supported by meta-analyses of patients with lower respiratory infections, including those treated in an intensive care unit. Serial chest X-rays can shorten symptom duration and reduce disease comorbidities76,77.
CT and MRI
Supplementary diagnostic testing for COVID-19 provides affirmation and monitoring of viral infection. Conventional CXR possesses sensitivity of nearly 60% for initial detection of COVID-19-related pulmonary disease80. These CXR abnormalities include bilateral lower zone and peripherally predominant consolidation and hazy opacities78. In addition, CT scans demonstrate a ‘reversed halo’ pattern and signs of septal thickening81. Distinctive CT images illustrate bilateral pulmonary parenchymal ground-glass and consolidated pulmonary opacities with occasionally rounded morphology and marginal lung dispersal (Fig. 4a). Lung engrossment with peripheral predominance is seen in individuals with both SARS-CoV and MERS-CoV infections. However, chest CT showing pulmonary ground-glass opacities and alliance is more indicative of SARS-CoV-2 infection76,82.
a, Comparison between lung tissue from an uninfected person (left) and from a person infected with SARS-CoV-2 (right) in a representative CT scan. In these images, ground-glass opacification (light hazy grey tissue) is seen due to inflammatory responses. ARDS results in fluid accumulation in affected lung tissue independent of cardiac dysfunction (non-cardiogenic pulmonary oedema). Viral infection causes lung injury, leading to loss of gas exchange, atelectasis and hypoxaemia. ARDS is associated with fibrinous organizing pneumonia and alveolar damage. SARS-CoV-2 causes epithelial infection and alveolar macrophage inflammation, and activation and secretion of a range of proinflammatory and chemotactic factors that lead to progressive lung tissue damage. b, A SARS-CoV-2-infected patient’s brain MRI scan image showing the brain regions typically involved in those who develop encephalitis or acute necrotizing encephalopathy. The rims of the lateral ventricles can illustrate contrast enhancement with meningeal involvements (red arrow). The medial temporal lobes (yellow arrows), including the hippocampi, may show hyperintense signals, indicating inflammation that may result from the ‘cytokine storm syndrome’, and hypointense signals that show haemorrhage. Other brain regions including the thalamus, cerebral white matter, brain stem and cerebellum can be involved.
Patients with negative RT–PCR tests for SARS-CoV-2 can present with abnormal chest CT scans and later be diagnosed with COVID-19 (ref. 77). Thus, emerging evidence supports the use of chest CT examination as a confirmatory test for COVID-19 disease when patients have negative RT–PCR tests but high clinical suspicion for SARS-CoV-2 infection. Chest CT scans could be used as a diagnostic tool for patients with negative RT–PCR screening, as an adjunctive test, in combination with repeated RT–PCR assays. Specifically, high-resolution chest CT is vital for confirmatory analysis and evaluation of disease severity in patients with suspected SARS-CoV-2 infection76. Numerous studies have scrutinized chest CT images of patients infected with SARS-CoV-2, considering that abnormalities may also be due to other causes of pneumonia, leading to false-positive results. In one study performed in TongJi Hospital, Wuhan, involving 1,014 patients who were examined with both chest CT and RT–PCR tests, 601 patients (59%) had positive RT–PCR results, and 888 (88%) had positive chest CT scans. While the sensitivity of chest CT scans for COVID-19 was 97%, on the basis of positive RT–PCR results, 75% (308 of 413 patients) had positive chest CT scans with negative RT–PCR results77. In advanced cases, SARS-CoV-2 infection can lead to extensive lung tissue damage with reduced oxygen uptake in infected people76,82. Although chest CT abnormalities may precede symptom onset in 44% of the patients with COVID-19, >90% of those with respiratory symptoms will have abnormal chest CT after symptom onset81. Furthermore, abnormalities on chest CT may be seen even in asymptomatic individuals with positive RT–PCR, as reported in 14 of 15 healthcare workers in one study83 and 54% of 82 asymptomatic passengers on the Diamond Princess cruise ship76,82. Despite the sensitivities of chest CT to detect lung abnormalities, the current recommendations from the major radiological societies are that chest CT should not be used for first-line screening of COVID-19, but should be used sparingly for hospitalized, symptomatic patients with specific clinical indications. Normal CT should not dissuade a patient from being quarantined or provided with other clinically indicated treatment when otherwise medically appropriate84. Most recently, a Consensus Statement from the Fleischner Society was generated by a multidisciplinary panel comprised principally of radiologists and pulmonologists from 10 countries with experience managing patients with COVID-19 across a spectrum of healthcare environments, evaluating the utility of imaging within three scenarios representing varying risk factors, community conditions and resource constraints85. On the basis of 14 key questions, corresponding to 11 decision points within the three scenarios and three additional clinical situations, the aggregated results yielded the following recommendations85 (see Table 1).
The immune response to SARS-CoV-2 leads to the release of cytokines and chemokines, frequently leaving inflammatory cells, which can be seen by CT (Fig. 4a) in the form of yellow discolouration. Figure 4b shows MRI scans of the brain of a patient infected with COVID-19, which provide much more detail of the pathologies in the soft tissue than CT. However, the American College of Radiology advises medical facilities to avoid performing MRI in COVID-19 patients. According to the American College of Radiology’s published guidelines, patients who are suspected of SARS-CoV-2 infection or have tested positive can be scanned by alternative imaging methods. Sanitizing MRI machines takes a long time and poses significant challenges. High-efficiency particulate air (HEPA) filter systems, typically used to increase air exchange, are not compatible with MRI. In case the ventilation examination is deemed clinically necessary, it is recommended that the potential risks of a patient having COVID-19 should be discussed with the referring physician and an alternative ventilation scan should be offered as per the hospital COVID-19 policies86.
Ultrasound
Ultrasonography of the lungs is also used to assess patients with COVID-19 (ref. 87). LUS does not appear to have specificity for identifying COVID-19 pneumonitis or pneumonia but is recommended for defining the area of infection. LUS may be beneficial in the early diagnosis of COVID-19 pneumonia as a cost-effective way to determine the localization of infection. The result of LUS is more sensitive than a CXR due to its excellent response to positive end-expiratory pressure (PEEP: pressure in the lungs above atmospheric pressure that exists at the end of expiration). LUS shows several features, such as lung consolidation in severe local disease. Similar to that found with CXR or chest CT, LUS in patients with COVID-19 infection found more prominent evidence of COVID-19 pathology in the posterior lower lung zones. In most cases, the infection progresses from the periphery to the centre of the lung tissues. Intensive care unit teams also use LUS findings of pulmonary oedema to therapeutically position the patients87.
Limitations of current diagnostic testing
Currently, a ‘clinical diagnosis’ of COVID-19 relies on a combination of chest CT and RT–PCR results. Outside a clinical setting, RT–PCR testing comprises the vast majority of surveillance testing done in the workplace or within schools. Due to the ubiquity of RT–PCR testing, it is important to examine the information this test offers to clinicians and policy makers alike. By identifying the shortcomings of this testing platform, future detection methods can improve upon the current model. Nucleic acid amplification tests may be problematic with poorly timed specimen collection, poor-quality specimen collection, the requirement for trained laboratory technicians and long wait times to generate the results. The gold standard RT–qPCR (quantitative PCR) is time consuming (4–6 h), not including the time to transport the specimens to the laboratory, which can take days. RT–PCR results also depend heavily on the type of sample taken: positive sampling rates vary widely between oropharyngeal swabs (32–48%), nasopharyngeal swabs (63%), bronchoalveolar lavage fluid (79–93%), sputum (72–76%) and stool (29%) (ref. 88). There could be shortages of the primers and other reagents required to run the tests89. In response to the limitations posed by RT–PCR testing, new platforms are actively being pursued. Research on antibody detection tests are ongoing, but limited. Many studies have had small cohorts and, given the urgency to share scientific knowledge in this unprecedented time, have rapidly published results that should be viewed with a critical lens. Current issues for immunodiagnostic approaches include a lack of specificity, which is linked to false-positive results from antigens that are well conserved among different CoV species and cross-reactions with autoantibodies in autoimmune diseases. Immunodiagnostic approaches are most viable 7–11 d after exposure and are therefore less useful in diagnosis of acute infection90. Outside the physical limitations of the testing platform, the information produced by both nucleic acid amplification testing and serology testing fails to capture important metrics of COVID-19, such as the duration for which an individual is contagious or the prevalence of certain haplotypes in a population. To these ends, metagenomic detection procedures used in tandem with nucleic acid amplification techniques may lead to new insights for clinicians and epidemiologists alike. Although RT–qPCR is the current standard for detection of nucleic acids, new methods, such as pulse-controlled amplification, are being evaluated. Pulse-controlled amplification does not require RNA extraction and can be carried out in 10 min with a small device91. In the future, S- and N-based immunodiagnostic platforms will work alongside nucleic acid amplification tests to increase detection sensitivity of COVID-19 at minimal cost92. Future efforts towards the development of novel diagnostic platforms may prove fruitful if the tests are accurate, specific and easy to run, produce results in a short time, and are cheap to mass produce (Box 2). Given both the strengths and limitations of current testing platforms and their singular output values, the information afforded by testing results must be carefully scrutinized before making decisions in clinical and non-clinical settings. It is also imperative to consider other strategies such as cost-effective mass pooling and metagenomic profiling to predict future outbreaks (Box 3).
Conclusions
The SARS-CoV-2 pandemic follows a troublesome trajectory. The health, humanitarian, social and economic policies adopted by countries can influence the speed and strength of the recovery. Currently, no medication is usually recommended to treat COVID-19, and no cure is accessible. The US FDA has authorized medicines previously approved for other disorders to be used now as recommended therapy for COVID-19. The earlier lack of accessibility for testing has hampered the infection control; however, testing of this novel virus is increasing quickly. Diagnostic testing for COVID-19 is vital in detection of the virus, understanding its epidemiology, case management and suppressing transmission. Universal operating procedures and harmonization of the available diagnostic assays are needed for faster screening approaches in the global fight against the pandemic. Similarly, academic scientists and biotechnologists are charged with the description of additional SARS-CoV-2 strains to improve the cluster-based specificity and sensitivity of antibody and antigen-based tests. Significantly, nanomaterial-based virus detection technology can help in the development of high-sensitivity, simple, scalable, rapid and cost-effective COVID-19 detection tests that supply on-demand diagnostic capability effectively in the pandemic. This Review offers a road map for diagnostic strategies in the context of disease transmission and prevention. It is a basic science guide to better appreciate COVID-19 diagnostic complexities and to effect improved disease-combating strategies.
References
Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 5, 536–544 (2020).
Wang, D. et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. J. Am. Med. Assoc. 323, 1061–1069 (2020).
Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens for COVID-19 (Centers for Disease Control and Prevention, 2020); https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html
Machhi, J. et al. The natural history, pathobiology, and clinical manifestations of SARS-CoV-2 infections. J. Neuroimmune Pharmacol. 15, 359–386 (2020).
Wadman, M., Couzin-Frankel, J., Kaiser, J. & Matacic, C. How does coronavirus kill? Clinicians trace a ferocious rampage through the body, from brain to toes. Science (17 April 2020); https://www.sciencemag.org/news/2020/04/how-does-coronavirus-kill-clinicians-trace-ferocious-rampage-through-body-brain-toes
Udugama, B. et al. Diagnosing COVID-19: the disease and tools for detection. ACS Nano 14, 3822–3835 (2020).
Liu, R. et al. Positive rate of RT–PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin. Chim. Acta 505, 172–175 (2020).
Research Use Only 2019-Novel Coronavirus (2019-nCoV) Real-time RT-PCR Primers and Probes (Centers for Disease Control and Prevention, 2020); https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html
Wang, B., Li, R., Lu, Z. & Huang, Y. Does comorbidity increase the risk of patients with COVID-19: evidence from meta-analysis. Aging 12, 6049–6057 (2020).
Loeffelholz, M. J. & Tang, Y. W. Laboratory diagnosis of emerging human coronavirus infections—the state of the art. Emerg. Microbes Infect. 9, 747–756 (2020).
Winichakoon, P. et al. Negative nasopharyngeal and oropharyngeal swabs do not rule out COVID-19. J. Clin. Microbiol. 58, e00297-20 (2020).
Pan, Y. et al. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J. Infect. 81, e28–e32 (2020).
Lin, D. et al. Evaluations of the serological test in the diagnosis of 2019 novel coronavirus (SARS-CoV-2) infections during the COVID-19 outbreak. Eur. J. Clin. Microbiol. Infect. Dis. 39, 2271–2277 (2020).
Lipsitch, M., Swerdlow, D. L. & Finelli, L. Defining the epidemiology of Covid-19—studies needed. N. Engl. J. Med. 382, 1194–1196 (2020).
Okba, N. M. A. et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg. Infect. Dis. 26, 1478–1488 (2020).
Shen, Z. et al. Genomic diversity of severe acute respiratory syndrome-coronavirus 2 in patients with coronavirus disease 2019. Clin. Infect. Dis. 71, 713–720 (2020).
Xiao, F. et al. Infectious SARS-CoV-2 in Feces of Patient with Severe COVID-19. Emerg. Infect. Dis. 26, 1920–1922 (2020).
Huang, C. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020).
Liu, R. et al. Viral load dynamics in sputum and nasopharyngeal swab in patients with COVID-19. J. Dent. Res. 99, 1239–1244 (2020).
Yongchen, Z. et al. Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID-19 patients. Emerg. Microbes Infect. 9, 833–836 (2020).
Wang, W. et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 323, 1843–1844 (2020).
Kim, Y. G. et al. Comparison between saliva and nasopharyngeal swab specimens for detection of respiratory viruses by multiplex reverse transcription-PCR. J. Clin. Microbiol. 55, 226–233 (2017).
Parolo, C., Escosura-Muñiz, A. & Merkoçi, A. Enhanced lateral flow immunoassay using gold nanoparticles loaded with enzymes. Biosens. Bioelectron. 40, 412–416 (2013).
Huang, C., Wen, T., Shi, F., Zeng, X. & Jiao, Y. Rapid detection of IgM antibodies against the SARS-CoV-2 virus via colloidal gold nanoparticle-based lateral-flow assay. ACS Omega 5, 12550–12556 (2020).
Bullard, J. et al. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin. Infect. Dis. 71, 2663–2666 (2020).
Laboratory Testing for Coronavirus Disease 2019 (COVID-19) in Suspected Human Cases: Interim Guidance (World Health Organization, 2 March 2020); https://apps.who.int/iris/handle/10665/331329
Smyrlaki, I. et al. Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-PCR. Nat. Commun. 11, 4812 (2020).
Shieh, W. J. et al. Immunohistochemical, in situ hybridization, and ultrastructural localization of SARS-associated coronavirus in lung of a fatal case of severe acute respiratory syndrome in Taiwan. Hum. Pathol. 36, 303–309 (2005).
Zhu, X. et al. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens. Bioelectron. 166, 112437 (2020).
Corman, V. M. et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro. Surveill. 25, 2000045 (2020).
Eigner, U. et al. Clinical evaluation of multiplex RT-PCR assays for the detection of influenza A/B and respiratory syncytial virus using a high throughput system. J. Virol. Methods 269, 49–54 (2019).
Zhang, W. et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 9, 386–389 (2020).
Moulahoum, H., Ghorbanizamani, F., Zihnioglu, F., Turhan, K. & Timur, S. How should diagnostic kits development adapt quickly in COVID 19-like pandemic models? Pros and cons of sensory platforms used in COVID-19 sensing. Talanta 222, 121534–121534 (2020).
Zhu, X. et al. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens. Bioelectron. 166, 112437 (2020).
Augustine, R. et al. Loop-Mediated Isothermal Amplification (LAMP): A rapid, sensitive, specific, and cost-effective point-of-care test for coronaviruses in the context of COVID-19 pandemic. Biology (Basel) 9, 182 (2020).
Mohamed, E. T., Bau, H. H. & Song, J. A single and two-stage, closed-tube, molecular test for the 2019 novel coronavirus (COVID-19) at home, clinic, and points of entry. Preprint at https://doi.org/10.26434/chemrxiv.11860137.v1 (2020).
Hong, S., SuganyaSamson, A. A. & MyongSong, J. Application of fluorescence resonance energy transfer to bioprinting. TrAC Trends Analyt. Chem. 122, 115749 (2020).
Chacón-Torres, J. C., Reinoso, C., Navas-León, D. G., Briceño, S. & González, G. Optimized and scalable synthesis of magnetic nanoparticles for RNA extraction in response to developing countries’ needs for the detection and control of SARS-CoV-2. Sci. Rep. 10, 19004 (2020).
Hildebrandt, N. et al. Energy transfer with semiconductor quantum dot bioconjugates: A versatile platform for biosensing, energy harvesting, and other developing applications. Chem. Rev. 117, 536–711 (2017).
Oh, E. et al. Inhibition assay of biomolecules based on fluorescence resonance energy transfer (FRET) between quantum dots and gold nanoparticles. J. Am. Chem. Soc. 127, 3270–3271 (2005).
Moitra, P., Alafeef, M., Dighe, K., Frieman, M. B. & Pan, D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano 14, 7617–7627 (2020).
Gorshkov, K. et al. Quantum dot-conjugated SARS-CoV-2 spike pseudo-virions enable tracking of angiotensin converting enzyme 2 binding and endocytosis. ACS Nano 14, 2234–12247 (2020).
Yan, S., Sun, H., Bu, X. & Wan, G. New strategy for COVID-19: An evolutionary role for RGD motif in SARS-CoV-2 and potential inhibitors for virus infection. Front. Pharmacol. 11, 912 (2020).
Farzin, L., Shamsipur, M., Samandari, L. & Sheibani, S. HIV biosensors for early diagnosis of infection: The intertwine of nanotechnology with sensing strategies. Talanta 206, 120201 (2020).
Talebian, S., Wallace, G. G., Schroeder, A. & Stellacci, F. Nanotechnology-based disinfectants and sensors for SARS-CoV-2. Nat. Nanotechnol. 15, 618–621 (2020).
Seo, G. et al. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano 14, 5135–5142 (2020).
Tymm, C., Zhou, J., Tadimety, A., Burklund, A. & Zhang, J. Scalable COVID-19 detection enabled by lab-on-chip biosensors. Cell Mol. Bioeng. 13, 1–17 (2020).
Ahmadivand, A. et al. Functionalized terahertz plasmonic metasensors: Femtomolar-level detection of SARS-CoV-2 spike proteins. Biosens. Bioelectron. 177, 112971 (2021).
Huang, L. et al. One-step rapid quantification of SARS-CoV-2 virus particles via low-cost nanoplasmonic sensors in generic microplate reader and point-of-care device. Biosens. Bioelectron. 171, 112685 (2020).
Yanik, A. A. et al. An optofluidic nanoplasmonic biosensor for direct detection of live viruses from biological media. Nano Lett. 10, 4962–4969 (2010).
Azzi, L. et al. Saliva is a reliable tool to detect SARS-CoV-2. J. Infect. 81, e45–e50 (2020).
Soler, M. et al. Multiplexed nanoplasmonic biosensor for one-step simultaneous detection of Chlamydia trachomatis and Neisseria gonorrhoeae in urine. Biosens. Bioelectron. 94, 560–567 (2017).
Dang, T. et al. Protein binding kinetics quantification via coupled plasmonic-photonic resonance nanosensors in generic microplate reader. Biosens. Bioelectron. 142, 111494 (2019).
Shan, B. et al. Multiplexed nanomaterial-based sensor array for detection of COVID-19 in exhaled breath. ACS Nano 14, 12125–12132 (2020).
Qiu, G. et al. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano 14, 12125–12132 (2020).
Hou, H. et al. Detection of IgM and IgG antibodies in patients with coronavirus disease 2019. Clin. Transl. Immunol. 9, e01136 (2020).
Padoan, A. et al. IgA-Ab response to spike glycoprotein of SARS-CoV-2 in patients with COVID-19: a longitudinal study. Clin. Chim. Acta 507, 164–166 (2020).
Long, Q. X. et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 26, 845–848 (2020).
Varadhachary, A. et al. Salivary anti-SARS-CoV-2 IgA as an accessible biomarker of mucosal immunity against COVID-19. Preprint at medRxiv https://doi.org/10.1101/2020.08.07.20170258 (2020).
Pisanic, N. et al. COVID-19 serology at population scale: SARS-CoV-2-specific antibody responses in saliva. J. Clin. Microbiol. 59, e02204–20 (2020).
Petherick, A. Developing antibody tests for SARS-CoV-2. Lancet 395, 1101–1102 (2020).
Wolters, F. et al. Multi-center evaluation of Cepheid Xpert® Xpress SARS-CoV-2 point-of-care test during the SARS-CoV-2 pandemic. J. Clin. Virol. 128, 104426 (2020).
Green, K., Graziadio, S., Turner, P., Fanshawe, T. & Allen, J. Molecular and antibody point-of-care tests to support the screening, diagnosis and monitoring of COVID-19. CEBM Research https://www.cebm.net/wp-content/uploads/2020/04/POCT-Covid19.pdf (2020).
EUA Authorized Serology Test Performance (US Food and Drug Administration, 2020); https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/eua-authorized-serology-test-performance
Kohmer, N., Westhaus, S., Rühl, C., Ciesek, S. & Rabenau, H. F. Brief clinical evaluation of six high-throughput SARS-CoV-2 IgG antibody assays. J. Clin. Virol. 129, 104480 (2020).
Advice on the Use of Point-of-Care Immunodiagnostic Tests for COVID-19 (World Health Organization, 2020); https://www.who.int/docs/default-source/coronaviruse/sb-2020-1-poc-immunodiagnostics-2020-04-08-e.pdf?sfvrsn=4c26ac39_2
Abbott launches COVID-19 antibody test. Abbott (27 April 2020); https://www.abbott.com/corpnewsroom/product-and-innovation/abbott-launches-covid-19-antibody-test.html
Long, Q. X. et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 26, 1200–1204 (2020).
Pfaffe, T., Cooper-White, J., Beyerlein, P., Kostner, K. & Punyadeera, C. Diagnostic potential of saliva: current state and future applications. Clin. Chem. 57, 675–687 (2011).
Punyadeera, C., Dimeski, G., Kostner, K., Beyerlein, P. & Cooper-White, J. One-step homogeneous C-reactive protein assay for saliva. J. Immunol. Methods 373, 19–25 (2011).
Verma, D., Garg, P. K. & Dubey, A. K. Insights into the human oral microbiome. Arch. Microbiol. 200, 525–540 (2018).
Wyllie, A. L. et al. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. N. Engl. J. Med. 383, 1283–1286 (2020).
Nobel, Y. R. et al. Gastrointestinal symptoms and COVID-19: case–control study from the United States. Gastroenterology 159, 373–375.e2 (2020).
Novel coronavirus (SARS-CoV-2): Discharge Criteria for Confirmed COVID-19 Cases—When is it Safe to Discharge COVID-19 Cases from the Hospital or End Home Isolation? (European Centre for Disease Prevention and Control, 2020); https://www.ecdc.europa.eu/sites/default/files/documents/COVID-19-Discharge-criteria.pdf
Dennis, B. An early warning system for coronavirus infections could be found in your toilet. The Washington Post (1 May 2020); https://www.washingtonpost.com/climate-environment/2020/05/01/coronavirus-sewage-wastewater/
Hosseiny, M., Kooraki, S., Gholamrezanezhad, A., Reddy, S. & Myers, L. Radiology perspective of coronavirus disease 2019 (COVID-19): lessons from severe acute respiratory syndrome and Middle East respiratory syndrome. Am. J. Roentgenol. 214, 1078–1082 (2020).
Ai, T. et al. Correlation of chest CT and RT–PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology 296, E32–E40 (2020).
Wong, H. Y. F. et al. Frequency and distribution of chest radiographic findings in patients positive for COVID-19. Radiology 296, E72–E78 (2020).
Dennie, C. et al. Canadian Association of Thoracic Radiology/Canadian Association of Radiologists consensus statement regarding chest imaging in suspected and confirmed COVID-19. Can. Assoc. Radiol. J. 71, 470–481 (2020).
Wang, Y. et al. Temporal changes of CT findings in 90 patients with COVID-19 pneumonia: a longitudinal study. Radiology 296, E55–E64 (2020).
Bernheim, A. et al. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology 295, 200463 (2020).
Zhou, Z. et al. Coronavirus disease 2019: initial chest CT findings. Eur. Radiol. 30, 4398–4406 (2020).
Shi, H. et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect. Dis. 20, 425–434 (2020).
Czawlytko, C., Hossain, R. & White, C. S. Covid-19 diagnostic imaging recommendations. Appl. Radiol. 49, 10–15 (2020).
Rubin, G. D. et al. The role of chest imaging in patient management during the COVID-19 pandemic: a multinational consensus statement from the Fleischner Society. Radiology 296, 172–180 (2020).
ACR Guidance on COVID-19 and MR Use (Americal College of Radiology, 2020); https://www.acr.org/Clinical-Resources/Radiology-Safety/MR-Safety/COVID-19-and-MR-Use
Poggiali, E. et al. Can lung US help critical care clinicians in the early diagnosis of novel coronavirus (COVID-19) pneumonia? Radiology 295, E6 (2020).
Xu, Y., Cheng, M., Chen, X. & Zhu, J. Current approach in laboratory testing for SARS-CoV-2. Int. J. Infect. Dis. 100, 7–9 (2020).
D'Cruz, R. J., Currier, A. W. & Sampson, V. B. Laboratory testing methods for novel severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2). Front. Cell Dev. Biol. 8, 468 (2020).
Abduljalil, J. M. Laboratory diagnosis of SARS-CoV-2: available approaches and limitations. New Microbes New Infect. 36, 100713 (2020).
Zwirglmaier, K. et al. Rapid detection of SARS-CoV-2 by pulse-controlled amplification (PCA). Preprint at medRxiv https://doi.org/10.1101/2020.07.29.20154104 (2020).
Infantino, M. et al. Serological assays for SARS-CoV-2 infectious disease: benefits, limitations and perspectives. Isr. Med. Assoc. J. 22, 203–210 (2020).
Li, Q. et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 382, 1199–1207 (2020).
Dara, M. & Talebzadeh, M. CRISPR/Cas as a potential diagnosis technique for COVID-19. Avicenna J. Med. Biotechnol. 12, 201–202 (2020).
Li, Y., Li, S., Wang, J. & Liu, G. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 37, 730–743 (2019).
Gootenberg, J. S. et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, 438–442 (2017).
Kellner, M. J., Koob, J. G., Gootenberg, J. S., Abudayyeh, O. O. & Zhang, F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 14, 2986–3012 (2019).
Ioannidis, J. P. A. The infection fatality rate of COVID-19 inferred from seroprevalence data. Bull. World Health Organ. 99, 19–33F (2021).
Ackerman, C. M. et al. Massively multiplexed nucleic acid detection using Cas13. Nature 582, 277–282 (2020).
Huang, Z. et al. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 164, 112316 (2020).
Chen, J. S. et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439 (2018).
Broughton, J. P. et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 38, 870–874 (2020).
GenMark Diagnostics Announces Submission of Emergency Use Authorization for its ePlex® SARS-CoV-2 Test (GenMarkD, 2020); https://www.genmarkdx.com/genmark-diagnostics-announces-submission-of-emergency-use-authorization-for-its-eplex-sars-cov-2-test/
The BioFire FilmArray Respiratory (RP & RP2) Panels (BioFire, 2020); https://www.biofiredx.com/products/the-filmarray-panels/filmarrayrp/
Meridian Bioscience Simplifies COVID-19 Sample Prep and Eliminates Dependence on Reagents in Short Supply (Meridian Biosciences, 2020); https://investor.meridianbioscience.com/news-releases/news-release-details/meridian-bioscience-simplifies-covid-19-sample-prep-and
Xpert® Xpress SARS-CoV-2 has received FDA Emergency Use Authorization (Cepheid, 2020); https://www.cepheid.com/coronavirus
Abbott’s Fast, $5, 15-Minute, Easy-to-Use COVID-19 Antigen Test Receives FDA Emergency Use Authorization; Mobile App Displays Test Results to Help Our Return to Daily Life; Ramping Production to 50 Million Tests a Month (Abbott, 2020).
Cesselli, D. et al. Implementation and validation of a pooling strategy for a sustainable screening campaign for the presence of SARS-CoV-2. Preprint at medRxiv https://doi.org/10.1101/2020.08.28.20174946 (2020).
Deckert, A., Bärnighausen, T. & Kyei, N. N. Simulation of pooled-sample analysis strategies for COVID-19 mass testing. Bull. World Health Organ. 98, 590–598 (2020).
Litvak, E., Tu, X. M. & Pagano, M. Screening for the presence of a disease by pooling sera samples. J. Am. Stat. Assoc. 89, 424–434 (1994).
Wuhan tested millions of people for COVID-19 in just days. Could US cities do the same? Livescience (28 May 2020); https://www.livescience.com/pooled-sampling-covid19-in-wuhan-and-us-cities.html
Interim Guidance for Use of Pooling Procedures in SARS-CoV-2 Diagnostic, Screening, and Surveillance Testing (Centers for Disease Control and Prevention, 2020); https://www.cdc.gov/coronavirus/2019-ncov/lab/pooling-procedures.html
Hogan, C. A., Sahoo, M. K. & Pinsky, B. A. Sample pooling as a strategy to detect community transmission of SARS-CoV-2. J. Am. Med. Assoc. 323, 1967–1969 (2020).
Sahajpal, N. S. et al. Proposal of RT–PCR-based mass population screening for severe acute respiratory syndrome coronavirus 2 (coronavirus disease 2019). J. Mol. Diagn. 22, 1294–1299 (2020).
Lim, K. L. et al. A novel strategy for community screening of SARS-CoV-2 (COVID-19): sample pooling method. PLoS ONE 15, e0238417 (2020).
Mandavilli, A. Federal officials turn to a new testing strategy as infections surge. The New York Times (1 July 2020); https://www.nytimes.com/2020/07/01/health/coronavirus-pooled-testing.html
Peddu, V. et al. Metagenomic analysis reveals clinical SARS-CoV-2 infection and bacterial or viral superinfection and colonization. Clin. Chem. 66, 966–972 (2020).
Mora, M. et al. Highly matching coronavirus-like short sequences can be retrieved from environmental metagenomes. Preprint at Research Square https://doi.org/10.21203/rs.3.rs-44155/v1 (2020).
Van Tan, L. et al. SARS-CoV-2 and co-infections detection in nasopharyngeal throat swabs of COVID-19 patients by metagenomics. J. Infect. 81, e175–e177 (2020).
Moore, S. C. et al. Amplicon based MinION sequencing of SARS-CoV-2 and metagenomic characterisation of nasopharyngeal swabs from patients with COVID-19. Preprint at medRxiv https://doi.org/10.1101/2020.03.05.20032011 (2020).
Acknowledgements
We thank D. Meigs, University of Nebraska Medical Center, for proofreading and providing feedback on the manuscript. This work was supported by the National Institutes of Health R01 MH121402-01A1, R01 MH121402P01, R01 AG043540, R01 AG043530, P01 DA028555, P30 MH062261, R01 MH115860, R01 NS034249, R01 NS036126, the Carol Swartz Emerging Neuroscience Fund and the Margaret R. Larson Professorship. C.P. receives support from Cancer Australia (APP1145657) and the Garnett Passé and Rodney Williams Foundation.
Author information
Authors and Affiliations
Contributions
B.D.K. and H.E.G. conceptualized, organized, designed, interpreted and facilitated the writing of the manuscript. B.D.K. conceptualized and then created each of the figures and figure legends. B.D.K. and J. Machhi edited, wrote and revised the text. B.D.K., H.E.G., J. Machhi, J.H., M.D.O., N.B., D.S., S.D., M.P., A.M.S., S.G., C.P. and L.C. wrote sections and edited the manuscript; B.D.K., J. Machhi, H.E.G., J. McMillan, W.R.B., B.E., C.B.G., M.H., S.P.M.R. and R.E. structured the Review and proofed and edited the text. All authors read and approved the final manuscript contents.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Materials thanks Jeroen van Kampen, Jianwei Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kevadiya, B.D., Machhi, J., Herskovitz, J. et al. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 20, 593–605 (2021). https://doi.org/10.1038/s41563-020-00906-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41563-020-00906-z
This article is cited by
-
The characteristics of microbiome in the upper respiratory tract of COVID-19 patients
BMC Microbiology (2024)
-
Advances in nanobiosensors during the COVID-19 pandemic and future perspectives for the post-COVID era
Nano Convergence (2024)
-
Biporous silica nanostructure-induced nanovortex in microfluidics for nucleic acid enrichment, isolation, and PCR-free detection
Nature Communications (2024)
-
A novel saliva-based miRNA profile to diagnose and predict oral cancer
International Journal of Oral Science (2024)
-
Impact of SARS-CoV-2 Vaccination or Infection on the Safety and Efficacy of Aesthetic Injections: A Systematic Review
Aesthetic Plastic Surgery (2024)