Abstract
The oriental river prawn, Macrobrachium nipponense, is an important commercial aquaculture resource in China. During breeding season, short ovary maturation cycles of female prawns cause multi-generation reunions in ponds and affect the growth of females representing individual miniaturization (known as autumn -propagation). These reproductive characteristics pose problems for in large - scale farming. To date, the molecular mechanisms of reproduction regulation of M. nipponense remain unclear. To address this issue, we performed transcriptome sequencing and gene expression analyses of eyestalk and cerebral ganglia of female M. nipponense during breeding and non-breeding seasons. Differentially expressed gene enrichment analysis results revealed several important reproduction related terms and signaling pathways, such as “photoreceptor activity”, “structural constituent of cuticle” and “G-protein coupled receptor activity”. The following six key genes from the transcriptome were predicted to mediate environmental factors regulating reproduction of M. nipponense: neuroparsin, neuropeptide F II, orcokinin II, crustacean cardioactive peptide, pigment-dispersing hormone 3 and tachykinin. These results will contribute to a better understanding of the molecular mechanisms of reproduction of oriental river prawns. Further detailed functional analyses of the candidate reproduction regulation related neuropeptides are needed to shed light on the mechanisms of reproduction of crustacean.
Similar content being viewed by others
Introduction
The oriental river prawn Macrobrachium nipponense (Decapoda, Palaemonidae), is widely distributed in Japan, Korea, Vietnam and Myanmar1,2. It is also an important commercial fishery resource for aquaculture in China with an annual cultured production of about 265,061 tons and an annual cultured output value of near 20 billion RMB in 20153,4. Prawn aquaculture in China has entered into a rapid development period and become a very important sector in Chinese agriculture. However, the reproductive characteristics of M. nipponense pose problems for large scale farming. During breeding season, especially in late April, female oriental river prawns enter into period of a rapid development, displaying a short ovary maturation cycle and embryo and larval development are accelerated as water temperature increases. In August -October, the newborn females can mate and lay eggs even though they hatched only 45 days and are no more than 3 cm long. This sexual precocity phenomenon, also known as autumn -propagation, is very prevalent in both wild and farmed prawns (Fig. 1). It causes multi-generation reunions in ponds, which block pure line construction. It also increases breeding density, which can affect oxygen concentration and lead to greater feed consumption, thereby impacting breeding. Autumn propagation also affects the growth of female prawns, resulting in individual miniaturization and subsequent low market value of the product5.
The mechanisms that regulate reproduction in crustaceans have become a hot research topic in recent years. Advances in understanding insulin-like peptides in crustaceans and the ability to manipulate them and their encoding transcripts (IAGs) have raised the possibility of sexually manipulating crustacean populations6. However, our understanding of the molecular regulatory mechanisms involved in crustacean reproduction is still far from complete. Eyestalk and cerebral ganglia are known to be major sites for production and secretion of many hormones involved in crustacean reproduction7,8. Previous studies showed that the X-organ-sinus gland complex system in the eyestalk and cerebral ganglia secretes many important neuropeptide hormones (e.g., crustacean hyperglycaemic hormone, molt inhibiting hormone, vitellogenesis inhibiting hormone and mandibular organ-inhibiting hormone), pigment regulatory peptides (e.g., pigment-dispersing hormone, red pigment-concentrating hormone) and neurotransmitters (e.g., serotonin, 5-HT and dopamine)9,10,11. In addition, transcriptome sequencing has led to the discovery of important reproduction-related genes12,13,14,15,16,17. As a consequence, fairly long lists of neuropeptides are available for several decapods18,19,20. Large-scale next-generation sequencing studies of mixed tissues (including the eyestalk), ovary, testis, and the androgenic gland from M. nipponense have been conducted to identify reproduction- and ovary development-related genes21,22,23,24, such as vitellogenin25, insulin-like androgenic gland hormone-binding protein26, sperm gelatinase27, gonadotropin-releasing hormone like receptor28 and heat shock proteins29. However, the transcriptomic data that come from independent eyestalk and cerebral ganglia of M. nipponense was seldom reported.
In this paper, we present the comparative transcriptomes of eyestalk and cerebral ganglia from female M. nipponense in breeding and non-breeding season. The objectives of this study were to: (1) construct eyestalk and cerebral ganglia transcriptomes of female M. nipponense during breeding and non-breeding seasons; (2) identify differentially expressed genes (DEGs) and pathways that may play important roles in sexual precocity in M. nipponense; and (3) gain insight into the regulatory mechanisms involved in crustacean reproduction. The results of this work will contribute to a better understanding of the molecular mechanisms of reproduction of oriental river prawns.
Methods
Prawn material
Healthy adult female oriental river prawns of one-year-old, each approximately 2.26 ± 0.59 g in wet weight, were obtained from Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences, Wuxi, China.
Approximately 50 non-breeding season female prawns were captured in January, 2015 and cultured in a re-circulating water aquarium system and fed with paludina twice per day at water temperature (10 °C) for 3 days before sample collection. Prawns were anesthetized under MS222 anesthesia to minimize suffering prior to dissections. Five prawns formed one biological replicate, and three biological replicates were performed for each sampling time. Fresh cerebral ganglia and eyestalk tissue from each prawn, named as non-breeding season cerebral ganglia and eyestalk (NBSB and NBSE), was quickly collected and immediately and stored at −80 °C until processed.
About 100 breeding season individuals with different ovarian stages were obtained in June, 2015 and maintained under the same condition at water temperature (24 °C) for 3 days before sample collection. The ovarian cycle of prawns was classified into five stages based on the previous results30 and ovary color observation: Stage I (undeveloped stage, oogonium proliferation, transparent), Stage II (developing stage, primary vitellogenesis, yellow), Stage III (nearly-ripe stage, secondary vitellogenesis, light green), Stage IV (ripe stage, vitellogenesis termination, dark green) and Stage V (spent stage, gray). Prawns were anesthetized under MS222 anesthesia to minimize suffering prior to dissections. One prawn each of five ovarian stages formed one biological replicate, and three biological replicates were performed for each sampling time. Fresh cerebral ganglia and eyestalk tissue from each prawn, named as breeding season cerebral ganglia and eyestalk (BSB and BSE), was quickly collected and immediately and stored at −80 °C until processed. For qRT-PCR validation, five individual prawns were sampled and pooled into one at each ovarian stage and fresh cerebral ganglia and eyestalk tissue from each prawn was quickly collected and immediately stored at −80 °C for RNA isolation. Our study does not involve endangered or protected species. This study was approved by the Institutional Animal Care and Use Ethics Committee of the Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences (Wuxi, China).
RNA extraction, sequencing library preparation and next-generation sequencing
Total RNA from eyestalks and brains of non-breeding and breeding season was isolated using RNAiso Plus Reagent (TaKaRa, Japan) according to the manufacturer’s protocols. The RNA was then treated with RNase free DNase I (Takara, Japan) to avoid DNA contamination. The RNA integrity and quantity were determined using an Agilent 2100 Bioanalyzer (Agilent, Shanghai, China) and a NanoDrop 1000 spectrophotometer (Thermo Scientific, USA).
Poly (A) mRNA was briefly isolated using oligo (dT) beads. Short mRNA fragments were used as templates to synthesize the first-strand cDNA with random hexamers. The second-strand cDNA was synthesized using buffer, dNTPs, RNase H and DNA polymerase I. Short fragments were purified with Takara’s PCR extraction kit (Takara, Japan). Sequencing adapters were ligated to short fragments and resolved by agarose gel electrophoresis. Proper fragments were selected and purified and subsequently PCR amplified to create the final cDNA library template. A mixed cDNA sample representing three individuals s of tissues was prepared and sequenced using an Illumina HiSeq™ 2500 with 150 bp pair-end reads produced.
De novo assembly and annotation functional of the transcriptome
All the raw reads were initially processed to obtain clean reads using the following three steps. (a) Reads with adaptor contamination were discarded; (b) reads with ambiguous sequences “N” larger than 5% were removed; (c) low quality reads that contained more than 20% Q < 20 bases were discarded31.
The clean reads were assembled into non-redundant transcripts using the Trinity, which has been developed specifically for the de novo assembly of transcriptomes using short reads32. First, clean reads with a certain length of overlap were combined to generate contigs. If there were multiple duplicated reads, the redundant duplicated reads were removed and only one read was retained for assembly. Then, the paired-end reads were realigned to contigs to obtain transcripts, which could identify different contigs in the same transcript and ensure the interval among these contigs. The TGICL program (http://compbio.dfci.harvard.edu/tgi/software/) was then used to delete redundant transcripts and further assemble all transcripts to form a single set of unigenes. Finally, all of the clean reads were pooled together and assembled to form the global transcriptome data of M. nipponense.
For annotation, the assembled unigenes were annotated by the public protein databases, including, Swissprot protein (http://www.expasy.ch/sprot), clusters of orthologous groups (COG; http://www.ncbi.nlm.nih.gov/), non-redundant (NR; http://www.ncbi.nlm.nih.gov) and the Kyoto encyclopedia of genes and genomes (KEGG; http://www.genome.jp/kegg) databases using BLAST (E-value < 10−5). The Getorf software was used to predict coding regions of the assembled unigenes33. Based on Nr annotation, BLAST2 GO program was used for GO analysis (http://www.geneontology.org/). Further, COG classification and signal pathway annotation of unigenes was performed by BLASTx searching against the COG database and KEGG database.
Differential expression analysis
The expression of all unigenes was estimated by calculating read density as ‘reads per kb per million reads’ (RPKM) using the RSEM program34. To identify the DEGs, FDR ≤ 0.001 and two-fold change (log2 Ratio) ≥ 1 or ≤−1 were set to be the threshold for judging the significance of gene expression differences35. The gene expression profiles were compared with each other, and then all DEGs in each comparison were carried on the GO functional and KEGG pathway enrichment analysis using GO database and KEGG database36. The DEGs mapped to the KEGG database were screened and analyzed by MapMan software.
The identification of the simple sequence repeats (SSRs) and single nucleotide polymorphism markers (SNPs)
Microsatellite identification software (http://pgrc.ipk-gatersleben.de/misa/) was used to identify SSR markers. The repeat thresholds for mono-, di-, tri-, tetra-, penta- and hexa- nucleotide motifs were set as 10, 6, 5, 5, 5 and 5, respectively. The maximum number of bases interrupting two SSRs in a compound microsatellite was set as 100. The identification of SNPs for each library used SOAPsnp (http://soap.genomics.org.cn/soapsnp.html)37.
Real-time quantitative RT-PCR verification
The DEGs results were validated using qRT-PCR. The qRT-PCR assays were performed with three replicates, and the actin gene was used as an internal control to normalize the expression level of the target genes. The specific primers were designed according to the unigene sequences using Primer 5.0 software (Table S1).
Amplifications were performed on a 96-well plate with a 25 μL reaction volume containing 1 μL cDNA (50ng), 10 μL SsoFastTM EvaGreen® Supermix (BIO-RAD, CA, USA), 0.5 μL 10 μM of primers, and 13 μL of ddH2O. The PCR temperature profile was 95 °C for 30 s followed by 40 cycles of 94 °C for 15 s, 58 °C for 20 s and 72 °C for 20 s, with a 0.5 °C/5 s incremental increase from 60 °C to 95 °C. The relative copy number of mRNA was calculated according to the 2− ∆∆CT comparative CT method38. Quantitative data were expressed as means ± SD. Two-side t test was used to compare expression levels. A probability level of 0.05 was used to indicate significance. All statistics were performed using SPSS Statistics 19.0.
Results
Sequencing and de novo assembly
A total of 300,308,758 (about 89.85 Gb) clean reads representing 89,847,943,292 clean nucleotides (nt) were produced. Each sample yielded more than 7 Gb of data (at least 20 million reads). The average Q30 percentage and GC content were 91.03% and 46.49%, respectively (Table 1).
De novo sequence assembly generated 118,366 transcripts with a total length over 120,087,867 bp. The mean transcript size was 1,014.55 bp, and the transcript N50 was 1,698 bp. Finally, 90,491 unigenes were obtained after combining the transcripts, with a total length of 73,722,132 bp. The mean unigene length was 814.69 bp, and the N50 was 1,091 bp. In these unigenes, 52.68% were 300–500 bp, 27.60% were 500–1,000 bp, 11.96% were 1,000–2,000 bp, and 7.76% were >2,000 bp in size. An overview of the assembly results is provided in Tables 2 and 3. The raw data were uploaded to the National Center for Biotechnology Information (NCBI, PRJNA339889).
Functional annotation
In total, 34,183 unigenes significantly matched a sequence at a cut-off E-value of 10−5 in at least one of the public databases, which included Cluster of Orthologous Groups (COG), Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), Swissprot and NCBI non-redundant (NR). Of these, 22,282 unigenes over 300 bp were annotated and 11,901 unigenes larger than 1000 bp could be matched to known proteins. In total 33,012 unigenes matched NCBI NR annotations, which contained more annotated genes than the other databases (Table S2). Species most represented in the BLASTx searches included Zootermopsis nevadensis (2858), Daphnia pulex (1638) and Stegodyphus mimosarum (1033), but “other” species was the largest group (22279) (Fig. S1).
To analyze the functions of these unigenes, GO assignments were made. 13,295 unigenes were assigned at least one GO term, and all GO terms were classified into three groups and further divided into 55 functional subgroups. Among biological processes, more transcripts were assigned to “metabolic process” (7,742) and “cellular process” (6,478) than to other terms. Among the “molecular functions”, “binding-related genes” (6,339) and “catalytic activity” (6,278) were the most enriched GO terms. Among “cellular components”, transcripts assigned to “cell part” (4,545), “cell” (4,507) and “organelle” (3,086) were the three most abundant (Fig. S2).
When the assembled unigenes were annotated by the COG database, 9,646 unigenes with significant homology were classified into 25 COG clusters. Among the functional clusters, “general function prediction only” was the largest cluster (2,705 unigenes) and “extracellular structures” (1unigene) was the smallest one (Fig. S3).
To analyze the involved signal pathways, the unigenes were annotated by the KEGG database. A total of 9,243 unigenes were assigned to 221 KEGG pathways. The largest pathway group was protein processing in the “Ribosome” (ko03010), which contained 832 genes. This was followed by protein processing in “endoplasmic reticulum” (ko04141) and “RNA transport” (ko03013), which contained 340 and 280 genes, respectively.
Development of simple sequence repeats (SSR) and single nucleotide polymorphism (SNP) markers
In total, 14,899 SSRs were identified. They were located in 17,844 unigenes greater than 1 Kb in size, and 3667 unigenes contained more than one SSR (Table S3). The largest fraction of SSRs identified consisted of mono-nucleotides (7953), followed by di-nucleotides (3382) and tri-nucleotides (3357).
We also detected SNPs in each library (Table S4). In breeding season group, we found that the number of heterozygous SNPs (Hete SNPs) was larger than the homozygous SNPs (Homo SNPs) in eyestalk transcriptomes, whereas the opposite was true in brain transcriptomes. However, in non-breeding season group, the number of Hete SNPs was much lower than that of the Homo SNP in both eyestalk and cerebral ganglia transcriptomes.
Global analysis of DEGs
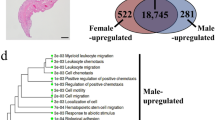
We calculated and compared expression levels of DEGs among the four groups to identify reproduction-related genes: cerebral ganglia of breeding season (BSB), eyestalks in breeding season (BSE), cerebral ganglia of non-breeding season (NBSB) and eyestalks of non-breeding season (NBSE). All DEGs with the absolute value of log2 Ratio ≥1 and the false discovery rate (FDR ≤ 0.001) are listed in Table S5. A total of 3,271 DEGs were detected for the NBSE and BSE, of which 2,136 and 1,135 unigenes were up-regulated and down-regulated respectively. For the NBSB and BSB comparison, 2,014 DEGs were detected, of which 1,318 and 696 unigenes were up-regulated and down-regulated respectively (Fig. 2).
Functional annotation of DEGs at a cut-off E-value of 10–5 was performed using COG, GO, KEGG, Swissprot and NCBI NR databases. Table S6 list the statistical results. GO functional annotation was performed to identify the biological function of DEGs (Fig. 3). Fewer of DEG unigenes had cellular component functions compared with the total group of unigenes. In addition, the annotations of functions of DEG unigene and all unigene for NBSE vs. BSE and NBSB vs. BSB comparisons were consistent with each other.
GO enrichment analysis was performed to identify significantly over-represented GO terms in the DEGs with a p-value < 0.05 and FDR < 0.01. The top 10 significant GO terms listed in Table S7. These terms included “photoreceptor activity”, “structural constituent of cuticle”, “G-protein coupled receptor activity”, “structural constituent of cytoskeleton”, “protein phosphatase type 2A regulator activity”, “iron ion binding”, “calcium ion binding”, “peptidase inhibitor activity”, “pigment binding” and “GTP binding”.
To better understand the reproduction- related pathways, DEGs also were analyzed using the KEGG database for pathway enrichment. We evaluated 20 most significant enriched pathways with the Q-value < 0.05 for the NBSE vs. BSE and NBSB vs. BSB comparisons (Fig. 4). For the eyestalk, the significantly enriched pathways between breeding season and non-breeding season included “phototransduction-fly” (Fig. 5), “glycolysis/gluconeogenesis” and “phagosome”. The most significantly enriched pathways for the cerebral ganglia between breeding season and non-breeding season were “drug metabolism– cytochrome P450” and “metabolism of xenobiotics by cytochrome P450”. Many other pathways, including “glycolysis/gluconeogenesis”, “phagosome”, “protein processing in endoplasmic reticulum”, “oxidative phosphorylation”, “glutathione metabolism” and “lysine biosynthesis”, were also enriched. Interestingly, when comparing eyestalk and cerebral ganglia of non-breeding season vs. breeding season, the DEGs were both concentrated most significantly in “phototransduction – fly”.
Significantly differentiated expressed genes that were identified by KEGG as involved in the “Phototransduction – fly” signaling pathway in NBSE vs. BSE. Notes: Red boxes indicate significantly increased expression. Black boxes indicate unchanged expression. Based on KEGG map0474536 (http://www.genome.jp/kegg-bin/show_pathway?ko04745).
Neuropeptides discovery by transcriptome mining and differentially expressed analyses
Further transcript characterization was performed for selected neuropeptides that were thought to play an important role in crustacean molting, feeding, metabolism, growth, and reproduction. Twenty-two transcripts encoding neuropeptide precursors were identified from the transcriptome data (Table S8).
The deduced peptides include red pigment concentrating hormone (RPCH), crustacean hyperglycemic hormone (CHH), molt inhibiting hormone (MIH), gonad inhibiting hormone (GIH), isoforms of allatostatin-B- I(AST-B-I), allatostatin B II I(AST-B- II), allatostatin-C(AST-C), neuropeptide F I(NPF-I), neuropeptide F II(NPF-II), short neuropeptide F (sNPF), sulfakinin, neuroparsin (NP), orcokinin II, crustacean cardioactive peptide (CCAP), eclosion hormone 2 (EH2), pigment dispersing hormone I (PDH-I), pigment-dispersing hormone 3(PDH3), tachykinin(TK), calcitonin-like diuretic hormone, corazonin preprohormone, crustacean female sex hormone and FLRFamide precursor protein B. Among these peptides, NP was found to be significantly up-regulated in the NBSE vs. BSE and down-regulated in the NBSB vs. BSB comparisons. NPF-II, orcokinin II, CCAP, PDH3 and TK were significantly down-regulated in the NBSE vs. BSE comparison. Sulfakinin were only various in the NBSE vs. BSE comparison.
Experimental validation
To validate the veracity and reliability of DEGs identified by RNA-Seq, we further examined relative expression levels of 10 genes of interest and quantified their relative expression folds between NBSE, BSE, NBSB and BSB groups by RT-qPCR. A total of 20 prawns from the NBSE, BSE, NBSB and BSB groups (n = 5 in each group) were used for each gene validation. The expression analysis was performed for selected genes belonging to the phototransduction, structural constituent of cuticle, and cytoskeleton biosynthesis pathways, including early cuticle protein 6, opsin, and calcification associated soluble matrix protein 2, tubulin beta-1 chain and myosin essential light chain. The qRT-PCR expression patterns of the 10 selected DEGs were in agreement with the results of RNA-Seq analysis (Fig. S4). In addition, a few transcripts encoding NP, CCAP, TK, PDH3 and NPFII showing differential expression, were also selected randomly from the transcriptome dataset and validated via real-time PCR analysis. A comparative analysis of all selected genes revealed similar expression pattern for real-time qPCR and RNA-seq data, illustrating the consistency of the results (Fig. 6).
Discussion
Transcriptomes of female M. nipponense eyestalk, and cerebral ganglia
Previous transcriptome analyses provide an insight into sex and reproduction of M. nipponense 23,24. In order to produce a comprehensive reference transcriptome, this study established a repertoire of annotated genes and DEGs from eyestalks and cerebral ganglia of female oriental river prawns during breeding and non-breeding seasons. From the four groups, 90,491 unigenes were assembled and investigated. Only 37.75% (34,183) of the unigenes significantly match with other known genes in the reference databases. BLAST results of the NR database showed that the majority of M. nipponense unigenes had the highest number of matches with the insect Z. nevadensis followed by the crustacean D. pulex. This result was similar to that reported for the M. nipponense ovary transcriptome but different from that reported for the M. rosenbergii eyestalk, CNS, and ovary transcriptome8,29. GO functional annotation and KEGG analysis revealed a great diversity of genes involved in various molecular functions.
Major biological associations explored by enrichment analysis
Photoreceptors
topGO term and pathway enrichment analyses were performed to explore GO terms and pathways in the DEGs related to ovarian maturation regulation. Comparison of proportions of GO and KEGG functional descriptions indicated that the enrichment trend was differed significantly between the DEGs and all genes. Both methods identified the same important signaling pathways: “photoreceptor activity” for GO term enrichment and “Phototransduction-fly” for KEGG enrichment. Light is primarily a source of energy for organisms, and it also can be acted as an environmental signal to control metabolic and reproductive processes39. In both birds and mammals, the photoperiodic pathways can regulate thyrotrophin secretion, which in turn regulates gonadotropin-releasing hormone secretion40,41. In invertebrates, the role of photoreceptors in insects also plays key roles in mates and other specific behaviors42. In this transcriptome data base, more than 300 elements belonging to the rhodopsin family were detected, and most of them were the part of DEGs designated as “photoreceptor activity” and “phototransduction-fly” term (opsin and amelanopsin). Similar results were also reported for the transcriptome of the crayfish Procambarus clarkii 43. In crustaceans, previous studies showed that the photoperiod has a strong induction effect on the gonad development44,45. However, no report about how light regulate reproduction are available. Based on results of this study, we speculate that a photoperiodic pathway may regulate in the reproduction crustaceans, as is true for birds and mammals. However, this hypothesis will require further studies to clarify the function of these rhodopsin family genes and elucidate the relationship between these photoreceptors and reproduction.
Cuticle proteins
Other inportant GO terms identified in this study are “structural constituent of cuticle” and “G-protein coupled receptor activity”. More than 110 cuticle related genes are included in the “structural constituent of cuticle” term including cuticle protein, early cuticle protein, strongly chitin-binding protein-1, and structural constituent of cuticle and calcified cuticle protein. Cuticular proteins are essential elements in cuticle occurrence and differentiation46,47. They play an indispensable role in insect cuticle integration, body shaping and other physiological process48,49,50. M. nipponense females cannot lay eggs unless they slough their old skin during breeding seasons which suggests a link between molting and reproduction. Molting is an important event for arthropod and ecdysteroids are multifunctional hormones in male and female arthropods regulate the construction of important parts and organs of insect body51. Previous studies have shown that ecdysteroid biosynthesis and its hormonal regulation are demanded for insect gonads52. Thus,Much needed be done to investigate the correlation between molting and reproduction in M. nipponense requires further study.
Reproduction regulation related neuropeptides identified by transcriptome mining
With the advancement of technology, such as genome and transcriptome mining and mass spectrometry, more than two dozen neuropeptide families have been characterized from various crustacean species53. Detail information about these peptides and their potential reproductive function of these neuropeptides is available for some insects54,55,56,57,58. In the current study, 22 transcripts encoding neuropeptide precursors were identified from the transcriptome data. However, only six neuropeptides (NP, CCAP, PDH, NPF–II, orcokinin–II, and TK) exhibited significant differences in the NBSE vs. BSE and NBSB vs. BSB comparisons. The other neuropeptides seemed not to react very strongly to seasonal alternation.
In this study, NP levels differed significantly varied in both the NBSE vs. BSE and NBSB vs. BSB comparisons. NPs, which had several isoforms, have been proved to play an important role in regulating reproduction of insects and crustaceans. They were first characterized from the brain of the locust, they were found to exhibit a vitellogenesis-inhibiting effect, preventing oocyte growth59,60,61. In crustaceans, the only exception of NP in Metapenaeus ensis that the gene silencing caused a significant decrease of vitellogenin expression level in both hepatopancreas and ovary62. Our results provide further evidence that NP might be involved in seasonal reproductive regulation. The main difference lies between breeding and non-breeding season was the variation of environmental factors. Light, temperature, and nutrition may be the most important factors affecting animal reproduction. Based on our results, we hypothesized that neuroparsin might mediate the way in which environmental factors regulating reproduction of M. nipponense.
Expression levels of the other five neuropeptides differed significantly between breeding season and non-breeding season neuropeptides only in the eyestalk. CCAP, which is found in neurons within the central nervous system of crustaceans and insects, was reported to play a role in accelerating heart contractions and hemolymph velocity58. It is also involved in modulating oviduct contraction in L. migratoria 63 and was significantly up-regulated in the late vitellogenic stage in S. paramamosain 64. PDH and RPCH are light adapting hormones that control circadian rhythm65,66. Several studies reported that PDH expression was maintained at high levels in the cerebral ganglia in the early vitellogenic stage but decreased significantly in the late vitellogenic stage in S. paramamosain 64,65. Neuropeptide F II (NPF-II), orcokinin II, and tachykinin (TK) have recently been reported by transcriptomes. In the cerebral ganglia transcriptome of S. paramamosain 66, differential expression levels of NPF-II, orcokinin II, and TK in the cerebral ganglia at different vitellogenic stages indicated that they may play a role in crustacean reproduction. However, the reproductive al function of these neuropeptides remains unknown and warrants further research. In the current study, significant expression differences of these neuropeptides were found only in the eyestalk, and phototransduction was the most important pathway. Based on this finding, we hypothesize that light might have a dominant effect on eyestalk neuropeptides secretion and that these five neuropeptides might mediate light regulating reproduction of M. nipponense. We currently are conducing further studies in our lab to test these hypotheses.
In summary, the comparative transcriptomes of eyestalk and cerebral ganglia from female M. nipponense during breeding and non-breeding seasons were evaluated in this study. We predicted major biological terms related to reproduction, such as photoreceptor activity and cuticle proteins, based on enrichment analysis. We also identified a series of peptides, including NP, CCAP, PDH, orcokinin II, TK, and NPF-II, which might mediate environmental factors that regulate reproduction of M. nipponense. These results provide insights into the mechanisms involved in reproduction of M. nipponense.
References
Yu, H. P. & Miyake, S. Five species of the genus Macrobrachium (Crustacea, Decapoda, Palaemonidae) from Taiwan. Ohmu. 3, 45–55 (1972).
Cai, Y. & Ng, P. K. L. The freshwater palaemonid prawns of Myanmar (Crustacea: Decapoda: Caridea). Hydrobiologia. 487, 59–83 (2002).
Fu, H. T., Jiang, S. F. & Xiong, Y. W. Current status and prospects of farming the giant river prawn (Macrobrachium rosenbergii) and the oriental river prawn (Macrobrachium nipponense) in China. Aquac. Res. 43, 993–998 (2012).
Bureau of Fishery, Ministry of Agriculture, P.R.C. Fisheries Economic Statistics. In: China Fishery Yearbook. Beijing, China Agricultural Press 236 (2016).
Qiao, H. et al. Characterization, expression, and function analysis of gonad-inhibiting hormone in Oriental River prawn, Macrobrachium nipponense and its induced expression by temperature. Comp Biochem Phys A. 185, 1–8 (2015).
Ventura, T. & Sagi, A. The insulin-like androgenic gland hormone in crustaceans: From a single gene silencing to a wide array of sexual manipulation-based biotechnologies. Biotechnol Adv. 30, 1543–1550 (2012).
Nagaraju, G. P. C. Reproductive regulators in decapod crustaceans: an overview. J Exp Biol. 214, 3–16 (2010).
Suwansa-ard, S. et al. In silico Neuropeptidome of Female Macrobrachium rosenbergii Based on Transcriptome and Peptide Mining of Eyestalk, Central Nervous System and Ovary. PLoS One. 10, e0123848 (2015).
Chan, S. M., Gu, P. L., Chu, K. H. & Tobe, S. S. Crustacean neuropeptide genes of the CHH/MIH/GIH family: implications from molecular studies. Gen Comp Endocrinol. 134, 214–219 (2003).
Treerattrakool, S., Boonchoy, C., Urtgam, S., Panyim, S. & Udomkit, A. Functional characterization of recombinant gonad-inhibiting hormone (GIH) and implication of antibody neutralization on induction of ovarian maturation in marine shrimp. Aquaculture. 428, 166–173 (2014).
Alfaro, J., Zúñiga, G. & Komen, J. Induction of ovarian maturation and spawning by combined treatment of serotonin and a dopamine antagonist, spiperone in Litopenaeus stylirostris and Litopenaeus vannamei. Aquaculture. 236, 511–522 (2004).
Gu, J. et al. Transcriptomic responses of corpuscle of stannius gland of Japanese eels (Anguilla japonica) to changes in water salinity. Sci. Rep. 5, 9836 (2015).
Luo, Y. et al. Transcriptomic profiling of differential responses to drought in two freshwater mussel species, the giant floater Pyganodon grandis and the pondhorn Uniomerus tetralasmus. PLoS One. 9, e89481 (2014).
Meng, X. et al. De novo characterization of Japanese scallop Mizuhopecten yessoensis transcriptome and analysis of its gene expression following cadmium exposure. PLoS One. 8, e64485 (2013).
Huang, S. et al. Transcriptomic variation of hepatopancreas reveals the energy metabolism and biological processes associated with molting in Chinese mitten crab, Eriocheir sinensis. Sci. Rep. 5, 14015 (2015).
Wang, W., Wu, X., Liu, Z., Zheng, H. & Cheng, Y. Insights into hepatopancreatic functions for nutrition metabolism and ovarian development in the crab Portunus trituberculatus: gene discovery in the comparative transcriptome of different hepatopancreas stages. PLoS One. 9, e84921 (2014).
Lv, J. et al. Transcriptome analysis of the Portunus trituberculatus: de novo assembly, growth-related gene identification and marker discovery. PLoS One. 9, e94055 (2014).
Suwansa-Ard, S. et al. In silico neuropeptidome of female Macrobrachium rosenbergii based on transcriptome and peptide mining of eyestalk, central nervous system and ovary. PLoS ONE. 10, e0123848 (2015).
Xu, Z. et al. Transcriptome profiling of the eyestalk of precocious juvenile Chinese mitten crab reveals putative neuropeptides and differentially expressed genes. Gene. 569, 280–286 (2015).
Veenstra, J. A. The power of next-generation sequencing as illustrated by the neuropeptidome of the crayfish Procambarus clarkii. Gen Comp Endocr. 224, 84–95 (2015).
Wu, P. et al. Gene discovery from an ovary cDNA library of riental river prawn Macrobrachium nipponense by ESTs annotation. Comp Biochem Phys D 4, 111–120 (2009).
Qiao, H. et al. Constructing and Random Sequencing Analysis of Normalized cDNA Library of Testis Tissue from Oriental River Prawn (Macrobrachium nipponense). Comp Biochem Phys D. 7, 268–276 (2012).
Jin, S. B. et al. Transcriptome analysis of androgenic gland for discovery of novel genes from the oriental river prawn, Macrobrachium nipponense, using Illumina Hiseq. 2000. PLOS ONE. 8, e76840 (2013).
Ma, K. et al. Transcriptome analysis of the oriental river prawn, Macrobrachium nipponense using 454 pyrosequencing for discovery of genes and markers. Plos One. 7, e39727 (2012).
Bai, H. K. et al. Molecular characterization and developmental expression of vitellogenin in the oriental river prawn Macrobrachium nipponense and the effects of RNA interference and eyestalk ablation on ovarian maturation. Gene. 562, 22–31 (2015).
Li, F. J. et al. Molecular characterization of insulin-like androgenic gland hormone-binding protein gene from the oriental river prawn Macrobrachium nipponense and investigation of its transcriptional relationship with the insulin-like androgenic gland hormone gene. Gen Comp Endocr. 216, 152–160 (2015).
Zhang, S. Y. et al. Molecular characterization, genomic organization, and expression analysis of sperm gelatinase gene during post-embryonic development in Macrobrachium nipponense. Turk J Fish Aquat Sc. 14, 689–696 (2014).
Du, Y. X., Ma, K. Y. & Qiu, F. Discovery of the genes in putative GnRH signaling pathway with focus on characterization of GnRH-like receptor transcripts in the brain and ovary of the oriental river prawn Macrobrachium nipponense. Aquaculture. 442, 1–11 (2015).
Jiang, H. X. et al. Insights into Sexual Precocity of Female Oriental River Prawn Macrobrachium nipponense through Transcriptome Analysis. PloS one. 11.6, e0157173 (2016).
Gao, X.G., Liu, H., Xu, J. N. & Cai, S.L. Study on Site ofVitellogenin Synthesis in the Freshwater Prawn Macrobrachium Nipponese. Biotechnology Bulletin, supplement 438–444 (2006).
Liu, T. et al. De novo assembly and characterization of transcriptome using Illumina paired-end sequencing and identification of CesA gene in ramie (Boehmeria nivea L. Gaud). BMC Genom. 14, 125 (2013).
Grabherr, M. G. et al. Regev A Fulllength transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29, 644–652 (2011).
Rice, P., Longden, I. & Bleasby AEMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277 (2000).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 12, 323 (2011).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
Kanehisa, M. et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, 353–361 (2017).
Li, R., Li, Y., Fang, X., Yang, H. & Wang, J. Kristiansen K SNP detection for massively parallel whole-genome resequencing. Genome Res. 19, 1124–1132 (2009).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C (T)) Method. Methods. 25, 402–408 (2001).
Rüdiger, W. & López-Figueroa, F. Photoreceptors in algae. Ann. Rev. Photochem. Photobiol. 55, 949–954 (1992).
Hanon, E. A. et al. Ancestral TSH mechanism signals summer in a photoperiodic mammal. Curr. Biol. 18, 1147–1152 (2008).
García-Fernández, J. M. et al. The hypothalamic photoreceptors regulating seasonal reproduction in birds: a prime role for VA opsin. Front Neuroendocr 37, 13–28 (2015).
Yan, S. et al. The expression of three opsin genes from the compound eye of Helicoverpa armigera (Lepidoptera: Noctuidae) is regulated by a circadian clock, light conditions and nutritional status. PloS one. 9, e111683 (2014).
Manfrin, C. et al. The eyestalk transcriptome of red swamp crayfish Procambarus clarkii. Gene. 557, 28–34 (2015).
Quackenbush, S. L. Lobster Reproduction: a Review. Crustaceana. 67, 82–94 (1994).
Matsuda, H., Takenouchi, T. & Yamakawa, T. Effects of photoperiod and temperature on ovarian development and spawning of the Japanese spiny lobster Panulirus japonicus. Aquaculture. 205, 385–398 (2002).
Delon, I. & Payre, F. Evolution of larval morphology in flies: get in shape with shavenbaby. Trends Genet. 20, 305–313 (2004).
Moussian, B., Schwarz, H., Bartoszewski, S. & Nusslein-Volhard, C. Involvement of chitin in exoskeleton morphogenesis in Drosophila melanogaster. J. Morphol. 264, 117–130 (2005).
Dittmer, N. T. et al. Proteomic and transcriptomic analyses of rigid and membranous cuticles and epidermis from the elytra and hindwings of the red flour beetle, Tribolium castaneum. J. Proteome Res. 11, 269–278 (2012).
Asano, T. et al. Identification of a cuticle protein with unique repeated motifs in the silkworm, Bombyx mori. Insect Biochem Molec. 43, 344–351 (2013).
Qiao, L. et al. Mutation of a cuticular protein, BmorCPR2, alters larvalbody shape and adaptability in silkworm. Bombyx mori. Genetics. 196, 1103–1115 (2014).
Teets, N. M. & Denlinger, D. L. Surviving in a frozen desert: environmental stress physiology of terrestrial Antarctic arthropods. J. Exp. Biol. 217, 84–93 (2014).
Brown, M., Sieglaff, D. & Rees, H. Gonadal ecdysteroidogenesis in Arthropoda: occurrence and regulation. Annu. Rev. Entomol. 54, 105–125 (2009).
Andrew, C. E. & Elizabeth, A. S. Crustacean neuropeptides. Cell. Mol. Life Sci. 67, 4135–4169 (2010).
Andrew, C. E. et al. Identification, tissue distribution and orexigenic activity of neuropeptide F (NPF) in penaeid shrimp. J Exp Biol. 214, 1386–1396 (2011).
Bissinger, B. W. et al. Synganglion transcriptome and developmental global gene expression in adult females of the American dog tick, Dermacentor variabilis (Acari: Ixodidae). Insect Mol. Biol. 20, 465–491 (2011).
Pieter, V. W., Senne, D., Sven, Z., Liesbeth, B. & Jozef, V. B. Regulation of feeding by neuropeptide F in the desert locust. Schistocerca gregaria. Insect Biochem. Mol. Biol. 43, 102–114 (2013).
Pieter, V. W., Niels, W. & Senne, D. Neuropeptide F regulates male reproductive processes in the desert locust. Schistocerca gregaria, Insect Biochem. Mol. Biol. 43, 252–259 (2013).
Estévez-Lao, T. Y., Boyce, D. S., Honegger, H. W. & Hillyer, J. F. Cardioacceleratory function of the neurohormone CCAP in the mosquito Anopheles gambiae. J Exp Biol. 216, 601–613 (2013).
Badisco, L. et al. RNA interference of insulin-related peptide and neuroparsins affects vitellogenesis in the desert locust Schistocerca gregaria. Peptides. 32(3), 573–580 (2011).
Marco, H. G., Anders, L. & Gäde, G. cDNA cloning and transcript distribution of two novel members of the neuroparsin peptide family in a hemipteran insect (Nezara viridula) and a decapod crustacean (Jasus lalandii). Peptides. 53, 97–105 (2014).
Brown, M. R. et al. Identification of a steroidogenic neurohormone in female mosquitoes. J. Biol. Chem. 273, 3967–3971 (1998).
Yang, S. P., He, J. G., Sun, C. B. & Chan, S. F. Characterization of the shrimp neuroparsin (MeNPLP): RNAi silencing resulted in inhibition of vitellogenesis. FEBS Open Bio. 4, 976–986 (2014).
Donini, A., Agricola, H. & Lange, A. B. Crustacean cardioactive peptide is a modulator of oviduct contractions in Locusta migratoria. J. Insect. Physiol. 47, 277–285 (2001).
Bao, C. C. et al. Neuropeptides in the cerebral ganglia of the mud crab, Scylla paramamosain: transcriptomic analysis and expression profiles during vitellogenesis. Sci Rep-UK. 5, 17055 (2015).
Huang, X., Ye, H., Huang, H., Yu, K. & Huang, Y. Two beta-pigment-dispersing hormone (β-PDH) isoforms in the mud crab, Scylla paramamosain: implication for regulation of ovarian maturation and a photoperiod-related daily rhythmicity. Anim Reprod Sci. 150, 139–147 (2014).
Fernlund, P. Structure of a light-adapting hormone from the shrimp, Pandalus borealis. Biochim. Biophys. Acta. 439, 17–25 (1976).
Acknowledgements
This research was supported by grants by Central Public-interest Scientific Institution Basal Research Fund CAFS (NO. 2015JBFM05); Central Public-interest Scientific Institution Basal Research Fund, CAFS(2016HY-ZD0402); China Agriculture Research System-47(CARS-47); National Natural Science Foundation of China (31572617); Science & Technology Supporting Program of Jiangsu Province (BE2016308), Fund of Independent Innovation of Agricultural Sciences of Jiangsu province (CX (15)1012–4); Fund of Three Innovations Engineering of Jiangsu Province (D2015–16); Science and Technology Development Fund of Wuxi(CLE02N1514). We also thank Genepioneer Biotechnologies for their help with the bioinformatics analyses.
Author information
Authors and Affiliations
Contributions
H.F. designed and supervised the study. H.Q. performed research and analyzed the transcriptome data. Y.X., S.J., W.Z., S.S., S.J. and Y.G. analyzed transcriptome data and prepared Tables and Figures; Y.W., F.L., D.S., and Y.W. performed qRT-PCR. H.Q. wrote the paper; which was read, reviewed and approved by all authors.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Qiao, H., Fu, H., Xiong, Y. et al. Molecular insights into reproduction regulation of female Oriental River prawns Macrobrachium nipponense through comparative transcriptomic analysis. Sci Rep 7, 12161 (2017). https://doi.org/10.1038/s41598-017-10439-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-10439-2
This article is cited by
-
Hepatopancreas transcriptome analyses provide new insights into the molecular regulatory mechanism of fast ovary maturation in Macrobrachium nipponense
BMC Genomics (2022)
-
Transcriptome analysis of five ovarian stages reveals gonad maturation in female Macrobrachium nipponense
BMC Genomics (2021)
-
Three neuroparsin genes from oriental river prawn, Macrobrachium nipponense, involved in ovary maturation
3 Biotech (2020)
-
Characterization of G-protein coupled receptors from the blackback land crab Gecarcinus lateralis Y organ transcriptome over the molt cycle
BMC Genomics (2019)
-
Molecular cloning, expression pattern analysis, and in situ hybridization of a Transformer-2 gene in the oriental freshwater prawn, Macrobrachium nipponense (de Haan, 1849)
3 Biotech (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.