Abstract
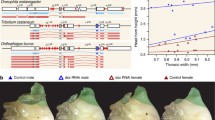
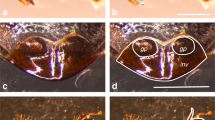
The origin, diversification, and secondary loss of sexually dimorphic characters are common in animal evolution. In some cases, structurally and functionally similar traits have evolved independently in multiple lineages. Prominent examples of such traits include the male-specific grasping structures that develop on the front legs of many dipteran insects. In this report, we describe the evolution and development of one of these structures, the male-specific “sex brush.” The sex brush is composed of densely packed, irregularly arranged modified bristles and is found in several distantly related lineages in the family Drosophilidae. Phylogenetic analysis using 250 genes from over 200 species provides modest support for a single origin of the sex brush followed by many secondary losses; however, independent origins of the sex brush cannot be ruled out completely. We show that sex brushes develop in very similar ways in all brush-bearing lineages. The dense packing of brush hairs is explained by the specification of bristle precursor cells at a near-maximum density permitted by the lateral inhibition mechanism, as well as by the reduced size of the surrounding epithelial cells. In contrast to the female and the ancestral male condition, where bristles are arranged in stereotypical, precisely spaced rows, cell migration does not contribute appreciably to the formation of the sex brush. The complex phylogenetic history of the sex brush can make it a valuable model for investigating coevolution of sex-specific morphology and mating behavior.





Similar content being viewed by others
Data availability
The datasets used in this study are included in the manuscript or will be made available in GenBank upon publication.
Code availability
Not applicable
References
Anisimova M, Gil M, Dufayard J-F, Dessimoz C, Gascuel O (2011) Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst Biol 60:685–699
Atallah J, Liu NH, Dennis P, Hon A, Godt D, Larsen EW (2009) Cell dynamics and developmental bias in the ontogeny of a complex sexually dimorphic trait in Drosophila melanogaster. Evol Dev 11:191–204
Atallah J, Watabe H, Kopp A (2012) Many ways to make a novel structure: a new mode of sex comb development in Drosophilidae. Evol Dev 14(6):476–83
Beaulieu JM, O’Meara BC, Donoghue MJ (2013) Identifying hidden rate changes in the evolution of a binary morphological character: the evolution of plant habit in campanulid angiosperms. Syst Biol 62:725–737
Da Lage J-L, Kergoat GJ, Maczkowiak F, Silvain J-F, Cariou M-L, Lachaise D (2007) A phylogeny of Drosophilidae using the Amyrel gene: questioning the Drosophila melanogaster species group boundaries. J Zool Syst Evol Res 45:47–63
Daugeron C, Plant A, Winkler I, Stark A, Baylac M (2011) Extreme male leg polymorphic asymmetry in a new empidine dance fly (Diptera: Empididae). Biol Let 7:11–14
dos Reis M, Yang Z (2011) Approximate likelihood calculation on a phylogeny for Bayesian estimation of divergence times. Mol Biol Evol 28:2161–2172
Drummond AJ, Suchard MA (2010) Bayesian random local clocks, or one rate to rule them all. BMC Biol 8:114
Eberhard WG (2001) The functional morphology of species-specific clasping structures on the front legs of male Archisepsis and Palaeosepsis flies (Diptera, Sepsidae). Zool J Linn Soc 133:335–368
Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinf 5:113
Felsenstein J (2005) Using the quantitative genetic threshold model for inferences between and within species. Philos Trans R Soc Lond B Biol Sci 360:1427–1434
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Höhna S, Landis MJ, Heath TA, Boussau B, Lartillot N, Moore BR, Huelsenbeck JP, Ronquist F (2016) RevBayes: Bayesian phylogenetic inference using graphical models and an interactive model-specification language. Syst Biol 65:726–736
Huber BA, Sinclair BJ, Schmitt M (2007) The evolution of asymmetric genitalia in spiders and insects. Biol Rev 82:647–698
Ingram KK, Laamanen T, Puniamoorthy N, Meier R (2008) Lack of morphological coevolution between male forelegs and female wings in Themira (Sepsidae : Diptera : Insecta). Biol J Lin Soc 93:227–238
Izumitani HF, Kusaka Y, Koshikawa S, Toda MJ, Katoh T (2016) Phylogeography of the subgenus Drosophila (Diptera: Drosophilidae): evolutionary history of faunal divergence between the old and the new worlds. PLoS ONE 11:e0160051
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Katoh TK, Zhang G, Toda MJ, Zhang WX, Gao JJ (2018) The Lordiphosa denticeps species group (Diptera: Drosophilidae) in China, with redescriptions of four known species and descriptions of nine new species. Zootaxa 4471:37–75
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C et al (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Kim BY, Wang JR, Miller DE, Barmina O, Delaney E, Thompson A, Comeault AA, Peede D, D’Agostino ER, Pelaez J et al (2021) Highly contiguous assemblies of 101 drosophilid genomes. Elife 10:e66405
Kopp A (2011) Drosophila sex combs as a model of evolutionary innovations. Evol Dev 13:504–522
Lanfear R, Calcott B, Ho SY, Guindon S (2012) Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol 29:1695–1701
Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B (2017) PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol 34:772–773
Lewis PO (2001) A likelihood approach to estimating phylogeny from discrete morphological character data. Syst Biol 50:913–925
Massey JH, Chung D, Siwanowicz I, Stern DL, Wittkopp P (2019) The yellow gene influences Drosophila male mating success through sex comb melanization. eLife 8:e49388
McAlpine JF (1981) Morphology and terminology - adults. In: McAlpine JF, Peterson BV, Shewell GE, Teskey HJ, Vockeroth JR, Wood DM (eds) Manual of Nearctic Diptera. Research Branch, Agriculture Canada, Ottawa, pp 9–63
Minh BQ, Nguyen MAT, von Haeseler A (2013) Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30:1188–1195
Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA (2018) Posterior summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst Biol 67:901–904
Revell LJ (2012) phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol 3:217–223
Rice G, Barmina O, Hu K, Kopp A (2018) Evolving doublesex expression correlates with the origin and diversification of male sexual ornaments in the Drosophila immigrans species group. Evol Dev 20:78–88
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542
Russo CA, Mello B, Frazao A, Voloch CM (2013) Phylogenetic analysis and a time tree for a large drosophilid data set (Diptera: Drosophilidae). Zool J Linn Soc 169:765–775
Simpson P (1990) Lateral inhibition and the development of the sensory bristles of the adult peripheral nervous system of Drosophila. Development 109:509–519
Sivinski J (1997) Ornaments in the Diptera. Florida Entomologist 80:142–164
Spieth HT (1952) Mating behavior within the genus Drosophila (Diptera). Bull Am Mus Nat Hist 99:395–474
Stark JB, O’Grady PM (2009) Morphological variation in the forelegs of the Hawaiian Drosophilidae. I. The AMC clade. J Morphol 271:86–103
Suvorov A, Kim BY, Wang J, Armstrong EE, Peede D, D’Agostino ERR, Price DK, Waddell P, Lang M, Courtier-Orgogozo V et al (2022) Widespread introgression across a phylogeny of 155 Drosophila genomes. Curr Biol 32:111-123.e5
Tanaka K, Barmina O, Kopp A (2009) Distinct developmental mechanisms underlie the evolutionary diversification of Drosophila sex combs. Proc Natl Acad Sci 106:4764–4769
Tsacas L (2002) Le nouveau complexe africain Drosophila loiciana et l’espèce apparentée D. matileana n. sp. (Diptera : Drosophilidae). Ann Soc Entomol Fr 38:57–70
Tsacas L, Chassagnard MT (1990) The species of the Genus Zaprionus with the fore femora bearing spines (Diptera, Drosophilidae). Annales De La Societe Entomologique De France 26:461–487
Tsacas L, Chassagnard M-T (2000) Drosophila loiciana, nouvelle espèce africaine et redescription de son epèce affine D. pruinosa Dudas (Diptera, Drosophilidae). Revue Française D’entomologie 22:213–222
Watts JC, Flynn A, Tenhumberg B, Hebets EA (2019) Contemporary sexual selection does not explain variation in male display traits among populations. Evolution 73:1927–1940
Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591
Yassin A, David JR (2010) Revision of the Afrotropical species of Zaprionus (Diptera, Drosophilidae), with descriptions of two new species and notes on internal reproductive structures and immature stages. ZooKeys 51:33–72
Yassin A, Araripe LO, Capy P, Da Lage JL, Klaczko LB, Maisonhaute C, Ogereau D, David JR (2008) Grafting the molecular phylogenetic tree with morphological branches to reconstruct the evolutionary history of the genus Zaprionus (Diptera: Drosophilidae). Mol Phylogenet Evol 47:903–915
Yassin A, Da Lage JL, David JR, Kondo M, Madi-Ravazzi L, Prigent SR, Toda MJ (2010) Polyphyly of the Zaprionus genus group (Diptera: Drosophilidae). Mol Phylogenet Evol 55:335–339
Acknowledgements
We thank the US Drosophila species stock center, Ehime University Drosophila stock center, J.-R. David, S. Prigent, and M. Watada for Drosophila strains and specimens; Developmental Studies Hybridoma Bank for antibodies; A. Comeault, A. Gloss, M. Lang, D. Matute, D. Miller, V. Orgogozo, and N. Whiteman for genome assemblies; M. Toda for advice on Zaprionus phylogeny; G. Rice for D. immigrans images; and A. Yassin for comments on the manuscript. We also thank the MCB Light Microscopy Imaging Facility, Department of Cell Biology and Human Anatomy EM Lab, and Materials Science and Engineering AMCaT Lab at UC Davis for equipment and imaging assistance, and Gwyneth Card and W. Ryan Williamson for use of the Photron camera. AK and OB are grateful to N. Gompel for allowing some of this work to be conducted in his lab while on a sabbatical visit, and to members of the Gompel lab for treating them as their labmates.
Funding
This work was supported by NIH grant 5R35GM122592 to AK, NIH grant F32 GM135998 to BYK, and by funds from Ludwig-Maximilians-Universität München to Nicolas Gompel, in whose lab some of the work was conducted. The Olympus FV1000 confocal used in this study was purchased using NIH Shared Instrumentation Grant 1S10RR019266-01.
Author information
Authors and Affiliations
Contributions
AK conceived the study. KT, OB, BYK, AK and JHM performed experiments and collected data. KT, BYK, AS, AT, and AK analyzed the data. KT and AK wrote the manuscript with contributions from AT, BYK, and AS.
Corresponding author
Ethics declarations
Ethics approval
Not applicable
Consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare no competing interests.
Additional information
Responsible Editor: Nico Posnien
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Movie 1 (MP4 1980 KB)
Supplementary Movie 2 (MP4 1394 KB)
Supplementary Movie 3 (MP4 1491 KB)
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tanaka, K., Barmina, O., Thompson, A. et al. Evolution and development of male-specific leg brushes in Drosophilidae. Dev Genes Evol 232, 89–102 (2022). https://doi.org/10.1007/s00427-022-00694-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-022-00694-3