Abstract
Background
As the dominant species of Stomatopoda, Oratosquilla oratoria has not been fully cultivated artificially, and the fishery production mainly depends on marine fishing. Due to the lack of stomatopod genome, the development of molecular breeding of mantis shrimps still lags behind.
Methods and results
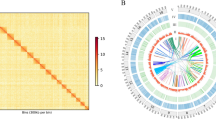
A survey analysis was performed to obtain the genome size, GC content and heterozygosity ratio in order to provide a fundation for subsequent whole-genome sequencing. The results showed that the estimated genome size of the O. oratoria was about 2.56 G, and the heterozygosity ratio was 1.81%, indicating that it is a complex genome. Then the sequencing data was preliminarily assembled with k-mer = 51 by SOAPdenovo software to obtain a genome size of 3.01G and GC content of 40.37%. According to ReapeatMasker and RepeatModerler analysis, the percentage of repeats in O. oratoria was 45.23% in the total genome, similar to 44% in Survey analysis. The MISA tool was used to analyze the simple sequence repeat (SSR) characteristics of genome sequences including Oratosquilla oratoria, Macrobrachium nipponense, Fenneropenaeus chinensis, Eriocheir japonica sinensis, Scylla paramamosain and Paralithodes platypus. All crustacean genomes showed similar SSRs characteristics, with the highest proportion of di-nucleotide repeat sequences. And AC/GT and AGG/CCT repeats were the main types of di-nucleotide and tri-nucleotide repeats in O. oratoria.
Conclusion
This study provided a reference for the genome assembly and annotation of the O. oratoria, and also provided a theoretical basis for the development of molecular markers of O. oratoria.





Similar content being viewed by others
Data availability
The datasets generated during the current study are available in the NCBI repository under BioProject: PRJNA935513.
References
Ahyong ST (2012) The Marine Fauna of New Zealand: Mantis Shrimps (Crustacea: Stomatopoda). Wellington: NIWA (National Institute of Water and Atmospheric Research Ltd). NIWA Biodiversity Memoir (ISSN 1174-0043; 125)
Zhao W, Yang QB, Chen X, Chen MQ, Wen WG (2019) A review of research on the biological properties and reproductive biology of some mantis shrimps. Marine Sci 43(4):105–114
Gutekunst J, Andriantsoa R, Falckenhayn C, Hanna K, Stein W (2018) Clonal genome evolution and rapid invasive spread of the marbled crayfish. Nat Ecol Evol 2(3):567–573
Veldsman WP, Ma KY, Hui JHL, Chan TF, Chu KH (2021) Comparative genomics of the coconut crab and other decapod crustaceans: exploring the molecular basis of terrestrial adaptation. BMC Genomics 22(1):1–5
Xu Z, Gao T, Xu Y, Li X, Tang J (2021) A chromosome-level reference genome of red swamp crayfish Procambarus clarkii provides insights into the gene families regarding growth or development in crustaceans. Genomics 113(5):3274–3284
Bachvaroff TR, Mcdonald RC, Plough LV, Chung JS (2021) Chromosome-level genome assembly of the blue crab Callinectes sapidus. G3 Genes|Genomes|Genetics 11(9):jkab212
Tang B, Zhang D, Li H, Jiang S, Zhang H, Xuan F, Ge B, Wang Z, Liu Y, Sha Z (2020) Chromosome-level genome assembly reveals the unique genome evolution of the swimming crab (Portunus trituberculatus). GigaScience 9(1):giz161
Xiong L, Wang Q, Qiu G (2012) large-scale isolation of microsatellites from chinese Mitten crab Eriocheir sinensis via a Solexa genomic survey. Int J Mole Sci 13(12):16333–16345
Zhang D, Ding G, Ge B, Zhang H, Tang B (2011) Development and characterization of microsatellite loci of Oratosquilla oratoria (Crustacea: Squillidae). Conserv Genet Resour 4(1):147–150
Dong X, Xing K, Sui Y, Liu H (2015) Genetic diversity of oratosqilla oratoria from four sea waters based on the mitochondrial coi gene sequences analysis. Marine Sci 39(7): 29–36
Cloonan N, Arr F, Kolle G, Bba G, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G (2008) Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nature methods 5(7):613–619
Kingsford C (2011) A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27(6):764–770
Ranallo-Benavidez T, Jaron K, Schatz M (2020) GenomeScope 2.0 and smudgeplot for reference-free profiling of polyploid genomes. Nat Commun 11(1):1432
Li R, Li Y, Kristiansen K, Wang J (2008) SOAP: short oligonucleotide alignment program. Bioinf (Oxford England) 24(5):713–714
Sebastian B, Thomas Thiel, Münch Uwe, Scholz Martin, Mascher,(2017) MISA-web: a web server for microsatellite prediction. Bioinformatics 33(16):2583–2585
Tarailo-Graovac M, Chen N (2009) Using RepeatMasker to identify repetitive elements in genomic sequences. Current protoc Bioinform 4:1–4
Robert C, Edgar (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Sudhir K, Glen S, Koichiro T (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biology Evol 33:1870–1874
Balakrishnan S, Gao S, Lercher MJ, Hu S, Chen WH (2019) Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res 47(W1):W270–W275
Wang Q, Ren X, Liu P, Li J, Lv J, Wang J, Zhang H, Wei W, Zhou Y, He Y et al (2022) Improved genome assembly of chinese shrimp (Fenneropenaeus chinensis) suggests adaptation to the environment during evolution and domestication. Mol Ecol Resour 22(1):334–344
Jin S, Bian C, Jiang S, Han K, Xiong Y, Zhang W, Shi C, Qiao H, Gao Z, Li R et al (2021) A chromosome-level genome assembly of the oriental river prawn Macrobrachium nipponense. GigaScience 10(1):giaa160
Tang B, Wang Z, Liu Q, Wang Z, Ren Y, Guo H, Qi T, Li Y, Zhang H, Jiang S et al (2021) Chromosome-level genome assembly of Paralithodes platypus provides insights into evolution and adaptation of king crabs. Mol Ecol Resour 21(2):511–525
Cui Z, Liu Y, Yuan J, Zhang X, Ventura T, Ma K, Sun S, Song C, Zhan D, Yang Y et al (2021) The chinese mitten crab genome provides insights into adaptive plasticity and developmental regulation. Nat Commun 12(1):2395
Zhao M, Wang W, Zhang F, Ma C, Liu Z, Yang M, Chen W, Li Q, Cui M, Jiang K et al (2021) A chromosome-level genome of the mud crab (Scylla paramamosain estampador) provides insights into the evolution of chemical and light perception in this crustacean. Mol Ecol Resour 21(4):1299–1317
Shi L, Yi S, Li Y (2018) Genome survey sequencing of red swamp crayfish Procambarus clarkii. Mol Biol Rep 45(5):799–806
Hamilton JP, Buell CR (2012) Advances in plant genome sequencing. Plant J 70(1):177–190
Shi MJ, Cheng YY, Zhang WT, Xia XQ (2016) The evolutionary mechanism of genome size. Chinese Sci Bull 61:3188–3195
López-Flores I, Garrido-Ramos M (2012) The repetitive DNA content of eukaryotic genomes. Genome Dyn 7:1–28
Kapitonov V, Jurka J (1999) Molecular paleontology of transposable elements from Arabidopsis thaliana. Genetica 107:27–37
Schug M, Wetterstrand KA, Gaudette MS, Lim RH, Hutter CM, Aquadro CF (1998) The distribution and frequency of microsatellite loci in Drosophila melanogaster. Mol Ecol 7:57–70
Edwards Y, Elgar G, Clark MS, Bishop MJ (1998) The identification and characterization of microsatellites in the compact genome of the japanese pufferfish, Fugu rubripes: perspectives in functional and comparative genomic analyses. J Mol Biol 278(4):843–854
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, Fitzhugh W (2001) Initial sequencing and analysis of the human genome. Nature 409(6822):860–921
Wang Z, Weber JL, Zhong G, Tanksley SD (1994) Survey of plant short tandem DNA repeats. Theor Appl Genet 88:1–6
Zhang J, Jibin Huang C (2016) Genome-wide functional analysis of SSR for an edible mushroom Pleurotus ostreatus. Gene 575(2):524–530
Labbé J, Murat C, Morin E, Tacon FL, Martin F (2011) Survey and analysis of simple sequence repeats in the Laccaria bicolor genome, with development of microsatellite markers. Curr Genet 57(2):75–88
Haydar K, Ying L, Wieland M (2005) Survey of simple sequence repeats in completed fungal genomes. Mol Biology Evol 22(3):639–649
Zhang Y, Lou FR, Han ZQ (2019) Development of microsatellite markers in Oratosquilla oratoria Transcriptome. J Zhejiang Ocean Univ (Natl Sci) 38(2):95–99
Acknowledgements
The work was funded by National Natural Science Foundation of China (32070526) and sponsored by “Qing Lan Project” & “333 Project”.
Funding
This study was supported by National Natural Science Foundation of China (32070526).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Methodology was performed by XS, and GW. Formal analysis, data curation and investigation were performed by JY, WY and JX. The original draft was written by GD and DZ. Writing-review and editing were performed by GW, XS and BT. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sun, X., Wang, G., Yang, J. et al. Whole genome evaluation analysis and preliminary Assembly of Oratosquilla oratoria (Stomatopoda: Squillidae). Mol Biol Rep 50, 4165–4173 (2023). https://doi.org/10.1007/s11033-023-08356-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08356-x