Occurrence of Terranova larval types (Nematoda: Anisakidae) in Australian marine fish with comments on their specific identities
- Published
- Accepted
- Received
- Academic Editor
- María Ángeles Esteban
- Subject Areas
- Aquaculture, Fisheries and Fish Science, Marine Biology, Parasitology, Taxonomy
- Keywords
- Anisakidae, Terranova, Taxonomy
- Copyright
- © 2016 Shamsi and Suthar
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2016. Occurrence of Terranova larval types (Nematoda: Anisakidae) in Australian marine fish with comments on their specific identities. PeerJ 4:e1722 https://doi.org/10.7717/peerj.1722
Abstract
Pseudoterranovosis is a well-known human disease caused by anisakid larvae belonging to the genus Pseudoterranova. Human infection occurs after consuming infected fish. Hence the presence of Pseudoterranova larvae in the flesh of the fish can cause serious losses and problems for the seafood, fishing and fisheries industries. The accurate identification of Pseudoterranova larvae in fish is important, but challenging because the larval stages of a number of different genera, including Pseudoterranova, Terranova and Pulchrascaris, look similar and cannot be differentiated from each other using morphological criteria, hence they are all referred to as Terranova larval type. Given that Terranova larval types in seafood are not necessarily Pseudoterranova and may not be dangerous, the aim of the present study was to investigate the occurrence of Terranova larval types in Australian marine fish and to determine their specific identity. A total of 137 fish belonging to 45 species were examined. Terranova larval types were found in 13 species, some of which were popular edible fish in Australia. The sequences of the first and second internal transcribed spacers (ITS-1 and ITS-2 respectively) of the Terranova larvae in the present study showed a high degree of similarity suggesting that they all belong to the same species. Due to the lack of a comparable sequence data of a well identified adult in the GenBank database the specific identity of Terranova larval type in the present study remains unknown. The sequence of the ITS regions of the Terranova larval type in the present study and those of Pseudoterranova spp. available in GenBank are significantly different, suggesting that larvae found in the present study do not belong to the genus Pseudoterranova, which is zoonotic. This study does not rule out the presence of Pseudoterranova larvae in Australian fish as Pseudoterranova decipiens E has been reported in adult form from seals in Antarctica and it is known that they have seasonal presence in Australian southern coasts. The genetic distinction of Terranova larval type in the present study from Pseudoterranova spp. along with the presence of more species of elasmobranchs in Australian waters (definitive hosts of Terranova spp. and Pulchrascaris spp.) than seals (definitive hosts of Pseudoterranova spp.) suggest that Terranova larval type in the present study belong to either genus Terranova or Pulchrascaris, which are not known to cause disease in humans. The present study provides essential information that could be helpful to identify Australian Terranova larval types in future studies. Examination and characterisation of further specimens, especially adults of Terranova and Pulchrascaris, is necessary to fully elucidate the identity of these larvae.
Introduction
Psudoterranovosis (Hochberg & Hamer, 2010), the seafood borne parasitic disease, caused by larvae of Pseudoterranova, is another form of anisakidosis, that has caused concern to human beings. The disease is most common in the United States followed by Japan and Europe (Hochberg & Hamer, 2010). With the increased popularity of eating raw or slightly cooked seafood dishes, the number of cases have increased globally (Chai, Darwin Murrell & Lymbery, 2005). The symptoms of the disease vary and may include nausea, severe epigastric pain and other abdominal discomforts, “tingling throat syndrome” from a worm crawling in the upper esophagus or oropharynx, cough and vomiting up live or dead worms (Margolis, 1977). The life cycle of the Pseudoterranova spp. includes crustaceans and fish as their intermediate hosts and marine mammals as their definitive hosts (Anderson, 2000). Human infection occurs after eating infected seafood, therefore the presence of Pseudoterranova larvae in the flesh of fish can cause serious losses and problems for fish and fisheries industry across the world. For example, up to 36 worms per fish have been reported in cod populations from Norwegian waters (Jensen, Andersen & Desclers, 1994) or Icelandic cod fillets provided by the industry have been reported to be infected with 2.5–17.6 worms per kg fillet (Hafsteinsson & Rizvi, 1987). It has been estimated that detection and removal of the larvae thought to be Pseudoterranova from the flesh of Atlantic cod (Gadus morhua) and other demersal species, and the resultant downgrading and discard of product, cause an annual loss of $50 million in Atlantic Canada (McClelland, 2002). This implies the need for detection and accurate identification of these larvae in fish. One of the challenges in diagnosing of parasitic diseases is the specific identification of larval stages of parasites. Larval stages of nematodes cannot be identified reliably using morphological characters alone. This is a consequence of the small size of larval stages and the lack of a sufficient number of characteristic features (Shamsi, Gasser & Beveridge, 2011). Molecular approaches have gained prominence for accurate identification of anisakids, irrespective of developmental stage and sex of the parasite, and for establishing systematic relationships (e.g., Orecchia et al., 1986). Several studies showed that ITS-1 and ITS-2 are useful genetic markers for specific identifications of nematodes irrespective of their developmental stage or sex and to study their life cycle (e.g., Shamsi, Gasser & Beveridge, 2011). However, this approach relies on presence of ITS sequences for well identified adults.
In several countries other than Australia, the ability to recognise and diagnose anisakidosis/pseudoterranovosis caused by these larvae has been improved, resulting in progress towards understanding its epidemiology and clinical manifestations of the disease. In Australia, however, little is known about the disease, the causative agent and its epidemiology. Australia is an increasingly multicultural country where seafood prepared in all its forms is very popular. A confirmed case of human anisakidosis was published recently by Shamsi & Butcher (2011) and several unpublished cases are on record (Shamsi, 2014). Therefore, there has been an increasing awareness of anisakidosis in humans and the presence of anisakid larval types in marine fish in Australia (Shamsi, 2014).
A review of the literature shows that Terranova larval types have been reported quite often in Australian marine fish (e.g., Cannon, 1977; Doupe et al., 2003; Lester, Barnes & Habib, 1985; Moore et al., 2011) but there is no information on the specific identity of Terranova larval types reported in Australia. The dilemma with Terranova larval types is that it could belong to any of three genera of anisakid nematodes, including Terranova, Pulchrascaris or Pseudoterranova, whose adult stages have been reported from Australian waters. Members of Terranova and Pulchrascaris become adult in elasmobranchs and are not known to cause harm to humans whereas Pseudoterranova spp. become adult in marine mammals and there are numerous publications about their pathogenicity and human health impacts. The larval stages of all these genera, i.e., Terranova, Pseudoterranova and Pulchrascaris are morphologically very similar. The typical characteristic of these larvae is the location of the excretory pore at the anterior end of the nematode, presence of a ventriculus without an appendix and having an intestinal caecum (Deardorff, 1987; Gibson & Colin, 1982). Therefore, distinction between larval stages of these genera based solely on morphology can be challenging. With recent increasing awareness about the presence of anisakid larvae in Australian fish as well as the presence of human cases in the country, knowing the specific identity of Terranova larval types becomes very important. In the last decade, molecular tools have provided the opportunity for specific identification of larval stages of parasites and there have been several works in the Americas, European countries and Antarctica on specific identification of Terranova larval types (Arizono et al., 2011; Paggi et al., 1991). Therefore, the aim of the present study is to employ a combined molecular and morphological approach to investigate the occurrence of Terranova larval types in Australian marine fish and to determine their specific identity.
Materials and Methods
Parasite collection
A total of 137 fish belonging to 45 species, Abudefduf whitleyi (n = 2), Aldrichetta forsteri (n = 1), Atherinomorus vaigiensis (n = 1), Caesio cuning (n = 8), Carangoides fulvoguttatus (n = 1), Caranx ignobilis (n = 2), C. melampygus (n = 1), Carcharias taurus (n = 1), Chaetodon aureofasciatus (n = 1), C. auriga (n = 1), C. flavirostris (n = 2), C. lineolatus (n = 1), C. melannotus (n = 1), Chaetodon sp (n = 1), Coryphaena hippurus (n = 1), Engraulis australis (n = 2), Epinephelus cyanopodus (n = 1), Grammatorcynus bicarinatus (n = 3), Haplophryne sp. (n = 1), Heniochus monoceros (n = 2), H. singularius (n = 1), Istiompax indica (n = 3), Kajikia audax (n = 3), Lutjanus argentimaculatus (n = 2), L. bohar (n = 1), L. carponotatus (n = 4), L. fulviflamma (n = 1), L. sebae (n = 4), Makaira mazara (n = 3), Mugil cephalus (n =5), Pastinachus sephen (n = 1), Platycephalus laevigatus (n = 8), Platycephalus sp. (n = 2), Pristipomoides multidens (n = 3), Rhombosolea tapirina (n = 3), Sardinops sagax neopilchardus (n = 8), Scomber australasicus (n = 11), Seriola hippos (n = 2), S. lalandi (n = 17), Siganus fuscescens (n = 1), S. punctatus (n = 1), Sillago flindersi (n = 13), Sphyraena novaehollandiae (n = 4), Taeniomembras microstomus (n = 1), and Thunnus albacares (n = 1) were examined for infection with anisakid larval types. Fish were collected off Australian coasts, including Queensland, New South Wales, Victoria, South Australia and Western Australia. No fish were caught or killed for the purpose of this study. All fish were either already euthanized as part of other research projects or were bought from fishermen in various fish markets.
| Abbreviation | Scientific name | Specimen/Accession no. | Reference | |
|---|---|---|---|---|
| ITS-1 | ITS-2 | |||
| A.brevispiculata | Anisakis brevispiculata | AY826719 | AY826719 | Nadler et al. (2005) |
| A.brevispiculata1 | Anisakis brevispiculata | PSW4-1 | PSW4-2 | Shamsi, Gasser & Beveridge (2012) |
| A.pegreffii | Anisakis pegreffii | FN391850 | FN556997 | Shamsi, Gasser & Beveridge (2012) |
| A.pegreffii1 | Anisakis pegreffii | FN391851 | FN556998 | Shamsi, Gasser & Beveridge (2012) |
| A.physeteris | Anisakis physeteris | AY826721 | AY826721 | Nadler et al. (2005) |
| A.physeteris1 | Anisakis physeteris | AY603530 | AY603530 | Kijewska et al. (2008) |
| A.simplexC | Anisakis simplex C | FN391883 | FN391884 | Shamsi, Gasser & Beveridge (2012) |
| A.simplexs.s. | Anisakis simplex sensu stricto | AJ225065 | AB196672 | Abe, Ohya & Yanagiguchi (2005) |
| A.TMTP | Larva of Anisakis sp. (TMTP) | AY260555 | AY260555 | Pontes et al. (2005) |
| A.typica | Anisakis typica | AY826724 | AY826724 | Nadler et al. (2005) |
| A.typica1 | Anisakis typica | FN391887 | FN391889 | Shamsi, Poupa & Justine (2015) |
| A.ziphidarum | Anisakis ziphidarum | AY826725 | AY826725 | Nadler et al. (2005) |
| C.bancrofti | Contracaecum bancrofti | EU839572 | FM177883 | Shamsi et al. (2009b) |
| C.bancrofti1 | Contracaecum bancrofti | EU839573 | FM177887 | Shamsi et al. (2009b) |
| C.eudyptulae | Contracaecum eudyptulae | FM177531 | FM177562 | Shamsi et al. (2009b) |
| C.margolisi | Contracaecum margolisi | AY821750 | AY821750 | Nadler et al. (2005) |
| C.microcephalum | Contracaecum microcephalum | FM177524 | FM177528 | Shamsi et al. (2009b) |
| C.multipapillatum | Contracaecum multipapillatum | AM940056 | AM940060 | Shamsi et al. (2008) |
| C.ogmorhini | Contracaecum ogmorhini sensu stricto | FM177542 | FM177547 | Shamsi et al. (2009b) |
| C.osculatumA | Contracaecum osculatum A | AJ250410 | AJ250419 | Zhu et al. (2000) |
| C.osculatumB | Contracaecum osculatum B | AJ250411 | AJ250420 | Zhu et al. (2000) |
| C.osculatumbaicalensis | Contracaecum osculatum baicalensis | AJ250415 | AJ250416 | Zhu et al. (2000) |
| C.osculatumC | Contracaecum osculatum C | AJ250412 | AJ250421 | Zhu et al. (2000) |
| C.osculatumD | Contracaecum osculatum D | AJ250413 | AJ250418 | Zhu et al. (2000) |
| C.osculatumE | Contracaecum osculatum E | AJ250414 | AJ250417 | Zhu et al. (2000) |
| C.radiatum | Contracaecum radiatum | AY603529 | AY603529 | Kijewska et al. (2002) |
| C.rudolphiiA | Contracaecum rudolphii A | AJ634782 | AY603535 | Li et al. (2005) |
| C.rudolphiiB | Contracaecum rudolphii B | AJ634783 | AJ634911 | Li et al. (2005) |
| C.rudolphiiD | Contracaecum rudolphii D | FM210253 | FM210267 | Shamsi et al. (2009a) |
| C.rudolphiiD1 | Contracaecum rudolphii D | FM210254 | FM210265 | Shamsi et al. (2009a) |
| C.rudolphiiE | Contracaecum rudolphii E | FM210257 | FM210269 | Shamsi et al. (2009a) |
| C.rudolphiiE1 | Contracaecum rudolphii E | FM210258 | FM210273 | Shamsi et al. (2009a) |
| C.septentrionale | Contracaecum septentrionale | AJ634784 | AJ634787 | Li et al. (2005) |
| C.variegatum | Contracaecum variegatum | FM177531 | FM177537 | Shamsi et al. (2009b) |
| Contracaecumn.sp. | Contracaecumpyripapillatum | AM940062 | AM940066 | Shamsi et al. (2009b) |
| H.aduncum1 | Hysterothylacium aduncum | AJ225068 | AJ225069 | Zhu et al., 1998) |
| H.aduncum2 | Hysterothylacium aduncum | AB277826 | AB277826 | Umehara et al. (2008) |
| H.auctum | Hysterothylacium auctum | AF115571 | AF115571 | Szostakowska et al. (2001) |
| H.III | Hysterothylacium larval type III | FN811721 | FN811678 | Shamsi, Gasser & Beveridge (2013) |
| H.III-1 | Hysterothylacium larval type III | FN811723 | FN811681 | Shamsi, Gasser & Beveridge (2013) |
| H.IVA | Hysterothylacium larval type IV Genotype A | FN811724 | FN811690 | Shamsi, Gasser & Beveridge (2013) |
| H.IVB | Hysterothylacium larval type IV Genotype B | FN811730 | FN811682 | Shamsi, Gasser & Beveridge (2013) |
| H.IVGA | Hysterothylacium larval type IV Genotype A | FN811729 | FN811690 | Shamsi, Gasser & Beveridge (2013) |
| H.IVGA1 | Hysterothylacium larval type IV Genotype A | FN811729 | FN811691 | Shamsi, Gasser & Beveridge (2013) |
| H.IVGA2 | Hysterothylacium larval type IV Genotype A | FN811729 | FN811692 | Shamsi, Gasser & Beveridge (2013) |
| H.IVGB | Hysterothylacium larval type IV Genotype B | FN811730 | FN811683 | Shamsi, Gasser & Beveridge (2013) |
| H.IVGB1 | Hysterothylacium larval type IV Genotype B | FN811731 | FN811684 | Shamsi, Gasser & Beveridge (2013) |
| H.IVGB2 | Hysterothylacium larval type IV Genotype B | FN811733 | FN811685 | Shamsi, Gasser & Beveridge (2013) |
| H.V | Hysterothylacium larval type V | FN811738 | FN811699 | Shamsi, Gasser & Beveridge (2013) |
| H.VI | Hysterothylacium larval type VI | FN811740 | FN811701 | Shamsi, Gasser & Beveridge (2013) |
| H.VII | Hysterothylacium larval type VII | FN811749 | FN811709 | Shamsi, Gasser & Beveridge (2013) |
| H.VIII | Hysterothylacium larval type VIII | FN811750 | FN811710 | Shamsi, Gasser & Beveridge (2013) |
| Heterakisgallinarum | Heterakis gallinarum | JQ995320 | JQ995320 | Jimenez et al. (2012) |
| P.azarasi | Pseudoterranova azarasi | AJ413973 | AJ413974 | Zhu et al. (2002) |
| P.bulbosa | Pseudoterranova bulbosa | AJ413970 | AJ413971 | Zhu et al. (2002) |
| P.cattani | Pseudoterranova cattani | AJ413982 | AJ413984 | Zhu et al. (2002) |
| P.decipiens | Pseudoterranova decipiens | AJ413979 | AJ413980 | Zhu et al. (2002) |
| P.decipiens1 | Pseudoterranova decipiens | AJ413979 | AJ413978 | Zhu et al. (2002) |
| R.acus | Raphidascaris acus | AY603537 | AY603537 | Kijewska et al. (2008) |
| Terranovasp. | Terranova sp. | LN795828 | LN795872 | The present study |
| Terranovasp.1 | Terranova sp. | LN795851 | LN795871 | The present study |
Dead fish were cut open and first examined for presence of larval nematodes in the surface of the internal organs and also for gross pathology. Then the gastro-intestinal tract from mouth to anus was examined for the presence of nematodes. All nematodes found were washed in physiological saline and then preserved in 70% ethanol. A small piece of the mid-body of each nematode was excised for molecular study, and the rest of the nematode were used for microscopy.
Morphological examination
The anterior and posterior parts of each nematode were cleared in lactophenol and examined under a light microscope. Terranova larvae were identified according to the identification key proposed by Cannon (1977) and were selected for description and further molecular analyses. Illustrations were made using a microscope equipped with camera lucida.
Molecular study
Genomic DNA (gDNA) was isolated from all individual larvae identified morphologically as Terranova larval type, by sodium dodecyl-sulphate/proteinase K treatment, column- purified (Wizard™ DNA Clean-Up; Promega, Madison, WI, USA) and eluted into 45 µl of water. PCR was used to amplify the ITS-1 and ITS-2 regions using primer sets SS1: 5′-GTTTCCGTAGGTGAACCTGCG-3′ (forward) and NC13R: 5′-GCTGCGTT CTTCATCGAT-3′ (reverse) for the former and SS2: 5′-TTGCAGACACATTGAGCACT-3′ (forward) and NC2: 5′-TTAGTTTCTTTTCCTCCGCT-3′ (reverse) for the latter region, and cycling conditions, initial 94 °C/5′, then 94 °C/30″, 55 °C/40″, 72 °C/40″ × 30 cycles, 72 °C/5′ extension and 4 °C (Shamsi & Butcher, 2011). An aliquot (4 µl) of each amplicon was examined on a 1.5% w/v agarose gel, stained with GelRed™ and photographed using a gel documentation system.
Representative samples based on host species and geographical locations were selected for sequencing. Sequences were aligned using the computer program ClustalX (Thompson et al., 1997) and then adjusted manually. Polymorphic sites were designated using International Union of Pure and Applied Chemistry (IUPAC) codes. Pair-wise comparisons of sequence differences (D) were determined using the formula D = 1 − (M∕L), where M is the number of alignment positions at which the two sequences have a base in common, and L is the total number of alignment positions over which the two sequences are compared (Chilton, Gasser & Beveridge, 1995).
Phylogenetic analysis of the nucleotide sequence data for combined ITS-1 and ITS-2 regions were conducted in PAUP 4.0. Table 1 shows details of the taxa used to build phylogenetic trees. Two tree-building methods, neighbour-joining and maximum parsimony were employed for phylogenetic analysis. The outgroup employed was Heterakis gallinarum (Nematoda: Heteakoidea; GenBank accession numbers JQ995320 and JQ995320 for ITS-1 and ITS-2, respectively).
| Taxonomically important morphological character | Measurement/description |
|---|---|
| Body length | 6.6 (3.0–9.0) |
| Body width | 0.24 (0.18–0.28) |
| Tooth | Present |
| Lips morphology | Inconspicuous |
| Distance of nerve ring from anterior end | 0.37 (0.22–0.72) |
| Location of excretory pore | At anterior end |
| Oesophagus length | 0.88 (0.4–1.14) |
| Ratio of oesophagus length to body length | 14.3 (9.5–26.5%) |
| Ventriculus length | 0.38 (0.24–0.54) |
| Intestinal caecum length | 0.71 (0.50–0.90) |
| Tail morphology | Strongly annulated, conical, tapering smoothly |
| Tail length | 0.13 (0.12–0.14) |
| Ratio of tail length to body length | 2.2% (1.3–4.0%) |
Results
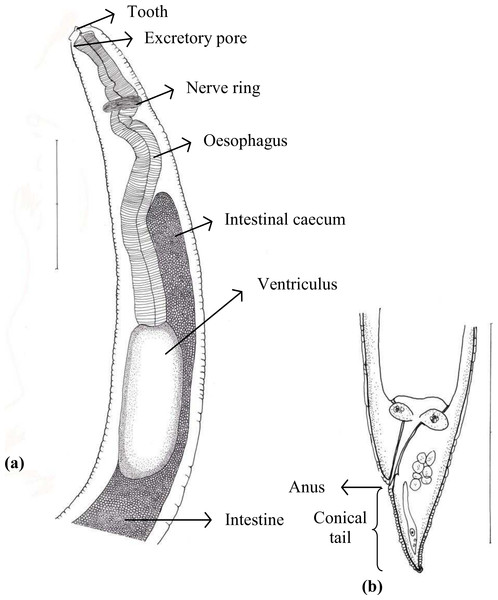
Of 45 species of fish examined in the present study, third stage Terranova type larvae (n = 93) were identified as type II based on the presence of intestinal caecum and ventriculus, absence of developed labia and ventricular appendix, and location of the excretory pore being at the anterior end (Fig. 1). Morphological description of these larvae was summarized in Table 2. Terranova type larvae were found in 13 species of fish collected from North-Eastern, Eastern and south eastern coasts of Australia. Material morphologically examined were 10 larvae in good condition from Caesio cuning (n = 3), Caranx ignobilis (n = 2), Grammatorcynus bicarinatus (n = 1), Lutjanus argentimaculatus (n = 3) and L. carponotatus (n = 1) from Heron Island, Queensland.
Figure 1: Diagram of Terranova larval type found in the present study indicating taxonomically important features (scale-bars = 0.3 mm).
| Taxa | Host common name | Host scientific name | Location | Reference |
|---|---|---|---|---|
| Terranova Leiper & Atkinson, 1914 | ||||
| T. amoyensis Fang & Luo 2006 | Red string ray | Dasyatis akajei | Taiwan Strait | Fang & Luo (2006) |
| T. antarctica (Leiper & Atkinson, 1914)a | Gummy shark | Mustelus antarcticus | Bay of Islands, New Zealand | Leiper & Atkinson (1914) |
| T. brevicapitata (Linton, 1901) | Tiger sharkk | Galeocerdo cuvier | Woods Hole, Massachusetts, USA | Deardorff (1987) |
| T. caballeroi Diaz-Ungria, 1967 | Porcupine river stingray | Potamotrygon hystrix | Delta of the Orinoco River, Venezuela | Diaz-Ungria (1967) |
| T. cephaloscyllii (Yamaguti, 1941) | Blotchy swell shark | Cephaloscyllium umbratile | Nagasaki, Japan | Yamaguti (1941) |
| T. circularis (Linstow, 1907) | Common sawfish | Pristis pristis | Cameroon | Bruce, Adlard & Cannon (1994) |
| T. crocodili (Taylor, 1924) | West African crocodile Malayan crocodile | Crocodylus sp Crocodylus johnstoni | Ghana Northern Australia; Queensland; Malaya | Sprent, (1979) |
| T. draschei (Stossich, 1896)b | Arapaima | Arapairna gigas | Rivers of northern South America | Bruce, Adlard & Cannon (1994) |
| T. galeocerdonis (Thwaite, 1927) | Tiger shark Scalloped hammerhead Smooth hammerhead Blacktai reef shark | Galeocerdo cuvier Sphyrna lewini S. zygaena Carcharinus amblyrhynchos | Twynams Paar, Ceylon; South Australia and Queensland, Australia; Natal, northern Brazil. | Bruce, Adlard & Cannon (1994) |
| T. ginglymostomae Olsen, 1952c | Nurse shark Spotted wobbegong Zebra shark | Ginglymostoma cirratum Orectolobus maculatusStegostoma fasciatum. | Tortugas, Florida, USA; off Queensland, Australia | Bruce, Adlard & Cannon (1994) |
| T. lanceolata (Molin 1860) | Black caiman American alligator | Melanosuchus niger Alligator mississippiensis | Brazil | Sprent (1979) |
| T. nidifex (Linton, 1900)d | Tiger shark | Galeocerdo tigrinus | Woods Hole, Massachusetts, USA | Deardorff (1987) |
| T. pristis (Baylis & Daubney, 1922) | Largetooth sawfish Snaggletooth shark Wallago | Pristis microdon (P. perotteti)Hemipristis elongatus Wallago attu | Ulubaria, India; Balgal, Queensland, eastern Australia | Bruce, Adlard & Cannon (1994) |
| T. petrovi Mozgovoi, 1950e | Shark | Raja longirostris | Kamchatka, USSR | Bruce, Adlard & Cannon (1994) |
| T. quadrata (Linstow, 1904) | The saltwater crocodile | Crocodilus porsus | Belgrade | Mozgovoi (1950) |
| T. rochalimai (Pereira, 1935)c | Shark | Scientific name was not mentioned in the original description | Brazil | Mozgovoi (1950) |
| T. scoliodontis (Baylis, 1931) | Shark | Scoliodon sp. | Cleveland Bay, Townsville, Australia | Bruce, Adlard & Cannon (1994) |
| T. secundum (Chandler, 1935)f | Largehead hairtail | Trichiurus lepturus. | Galveston Bay, Texas, USA; La Paloma, Uraguay | Chandler (1935) |
| T. serrata (Drasche, 1896)b | Arapaima | Arapaima gigas | Rivers of northern South America | Bruce, Adlard & Cannon (1994) |
| Terranova trichiuri (Chandler, 1935)g | Indian threadfin | Polydactylus indicus Trichiurus lepturus | Galveston Bay, Texas, USA; Khulna, Pakistan | Bruce, Adlard & Cannon (1994) |
| Pulchrascaris Vicente and dos Santos, 1972 | ||||
| P. caballeroi Vicente and dos Santos, 1972 | Angelshark | Squatina squatinah | Rio de Janeiro, Brazil | Bruce, Adlard & Cannon (1994) |
| P. chiloscyllii (Johnston and Mawson, 1951) | Brownbanded bambooshark Blacktip reef shark Gummy shark Scalloped hammerhead Smooth hammerhead Whitetip reef shark | Chiloscyllium punctatumCarcharinus melanopterus Mustelus antarcticus Sphyrna lewini S. zygaenaTriaenenodon obesus | Halfway Island, Australia; Hawaii, Alabama, USA; South Africa | Bruce, Adlard & Cannon (1994) |
| P. secunda (Chandler, 1935) | Largehead hairtail | Trichiurus lepturus. | Galveston Bay, Texas, USA; La Paloma, Uraguay | Bruce, Adlard & Cannon (1994) |
| Pseudoterranova Mozgovoi, 1951 | ||||
| Pseudoterranova azarasi (Yamaguti & Arima, 1942) | Steller’s sea lion Californian sea lion Harbor seal Bearded seal | Eumetopias jubatus Zalophus californianusPhoca vitulina richardsii Erignathus barbatus | Japanese and Sakhalinese waters of the North Pacific Ocean | Mattiucci & Nascetti (2008) |
| P. bulbosa (Cobb, 1888) | Bearded seal | Erignathus barbatus | Barents and Norwegian Seas, the Canadian Atlantic and the Sea of Japan, | Mattiucci & Nascetti (2008) |
| P. cattani George-Nascimento and Urrutia, 2000 | South American sea lion | Otaria byronia | South-East Pacific, Chilean coast | Mattiucci & Nascetti (2008) |
| P. decipiens (Krabbe, 1868) (sensu stricto) | Californian sea lion Harbor seal Harbor seal Grey seal Hooded seal Norhern elephant seal | Zalophus californianus Phoca vitulina richardsii Phoca vituline Halichoerus grypus Cystophora cristataMirounga angustirostris | North-East and North-West Atlantic | Mattiucci & Nascetti (2008) |
| P. krabbei Paggi, Mattiucci et al., 2000 | Harbor seal Grey seal | Phoca vituline Halichoerus grypus | North-East Atlantic; Faeroe Islands | Mattiucci & Nascetti (2008) |
| P. decipiens E of Bullini et al., 1997 | Antarctic Weddell seal | Leptonychotes weddellii | Antarctica | Mattiucci & Nascetti (2008) |
Notes:
A total of 93 specimens from various fishes, including Abudefduf whitleyi, Caesio cuning, Carangoides fulvoguttatus, Caranx ignobilis, Caranx melampygus, Epinephelus cyanopodus, Grammatorcynus bicarinatus, Lutjanus argentimaculatus, L. bohar, L. carponotatus and L. fulviflamma and Scomber australasicus were subjected to PCR amplification. Based on the species of hosts and their geographical locations, 25 and 21 specimens were selected and sequenced for ITS-1 and ITS-2 respectively.
The length of the ITS-1 was 437 bp except for two specimens which were 436 bp long. The difference in length was due to a gap at alignment position 20 in the latter specimens (Fig. 2). Also, sequence polymorphism was detected at alignment position 426 in one specimen (Fig. 2). Sequence variation in the ITS-1 among specimens was 0–0.4% and the G + C content was 47.6–47.9%. The length of the ITS-2 was 252 bp. Sequence polymorphism was detected at alignment position 22 in two specimens. Sequence variation among individuals was 0–0.4% and the G + C content was 46.4–46.8%. ITS-1 and ITS-2 sequences of Terranova larval type found in the present study were almost identical among all larvae.
Figure 2: Alignment of the sequences of the ITS-1 and ITS-2 regions of Terranova larval type II of Cannon, 1977c found in the present study.
The left column indicates the GenBank accession number of specimens. Numbers to the right of alignment indicate the alignment position. Polymorphic sites were designated using IUPAC codes.Discussion
Previously, Cannon (1977) described two distinct Terranova larval types, I and II, in Queensland waters which were later reported by other authors from other parts of Australia (e.g., Doupe et al., 2003; Moore et al., 2011). According to Cannon (1977), the main difference between larval types I and II is the ratio of intestinal caecum to ventriculus being 1:1 in the former and 2:1 in the latter morphotype. Based on the similarity in the ratio of intestinal caecum to ventriculus and considering the geographical location of larvae and matching it with presence of adult nematodes, he suggested Terranova larval type I in his study could be Terranova chiloscyiti and Terranova larval type II could be T. galeocerdonis or T. scoliodontis. Although some species within Pseudoterranova (e.g., P. cattani) have the same ratio of intestinal caecum to ventriculus and although Pulchrascaris has been reported from the same general location (Table 3), the possibility of these larvae being Pulchrascaris spp. or Pseudoterranova spp. was not discussed in Cannon’s work. In addition, assigning larval type to adults based on the ratio of intestinal caecum to ventriculus has been considered to be unreliable. Huizinga (1967) showed that the length of the intestinal caecum is shorter in smaller/younger larvae and increases as the larvae grow in length. This can affect the ratio of intestinal caecum to other organs, such as ventricular appendix or ventriculus. As a result the specific identity of Terranova larval types remains unknown. For the same reasons, despite of morphological resemblance between Terranova larval type in the present study and those described by Cannon (1977) there is no certainty that they are genetically similar or belong to the same species due to lack of comparable molecular data for Cannon’s specimens.
In an attempt to specifically identify Terranova larval type in the present study, we genetically characterised all Terranova larval type found in the present study from broad geographical region as well as a broad variety of fish species, based on their ITS-1 and ITS-2 sequences followed by phylogenetic analyses.
The nucleotide variation within Terranova larval type in the present study was very low (0–0.4% for both ITS-1 and ITS-2), and was within the range for nucleotide variation (0–0.2% and 0–0.4% for ITS-1 and ITS-2 respectively) calculated for members of the same species in the family Anisakidae (Shamsi et al., 2009b). This suggests they all should be the same genotype/species.
To reveal the specific identity of the Terranova larval type found in the present study comparable ITS sequences from well identified adults must be available. To date, there is no such sequence in the GenBank database. Among reliably identified species whose ITS-1 and ITS-2 sequences were available in the GenBank database, there was no identical or highly similar sequence to ITS-1 and ITS-2 sequences found in the present study. Alignment of ITS-1 and ITS-2 sequences of Terranova larval type in the present study with those available in GenBank database did not result in finding identical or highly similar sequences. Although the closest ITS sequences in the GenBank database belonged to Pseudoterranova azarasi, P. bulbosa, P. cattani and P. decipiens sensu strict the nucleotide difference between ITS sequences of the larvae in the present study and those of Pseudoterranova spp. in the GenBank was too great (38.9–39.8% and 46.7–48.4% for ITS-1 and ITS-2, respectively) to be considered within the genus Pseudoterranova (Table 4). The distinction between Terranova larval type in the present study and Pseudoterranova spp. was also supported by phylogenetic analyses (Fig. 3).
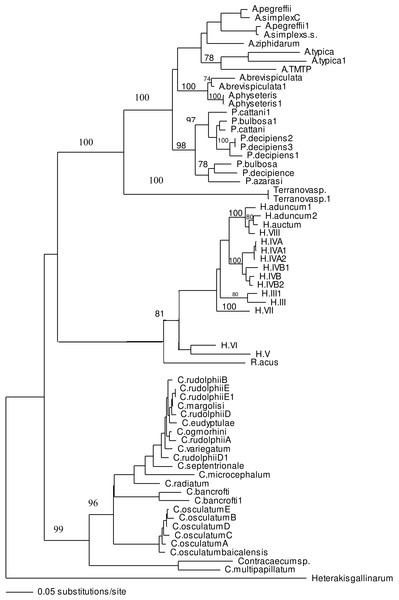
ITS-1 and ITS-2 sequences of well identified closely related taxa were selected to build the phylogenetic tree to investigate the association of larvae in the present study with other taxa within family Anisakidae. Both neighbour joining and maximum parsimony (the latter is not shown) trees had similar profile and grouping of taxa were the same among both trees. In the neighbour joining phylogenetic tree (Fig. 3), Terranova larval type found in the present study were resolved as a distinct clade with strong bootstrap support of 100%. None of the anisakid species (Pseudoterranova spp., Anisakis spp. and Contracaecum spp. becoming adult in marine mammals) with similar morphology to Terranova spp. (i.e., having excretory pore opened at the base of the labia) were grouped in the same clade as Terranova larval type found in the present study. Closely related species becoming adult in teleost fishes (Hysterothylacium spp. and Raphidascaris acus) were also included in the phylogentic tree, although the excretory system in this group has a different feature to Terranova spp. These anisakids also resolved as a distinct clade to Terranova larval type.
Figure 3: Phylogenetic analysis of the combined ITS-1 and ITS-2 sequence data for members of the Anisakidae with Heterakis gallinarum as outgroup, using the neighbour-joining method.
Bootstrap support values are indicated. See Table 1 for detailed abbreviations. Note that Terranovasp and Terranovasp1 both belong to the same taxon and only different in polymorphic sites as shown in Fig. 2. They are representative of 93 Terranova larval type examined in the present study.In both phylogenetic trees produced in the present study based on the combined ITS-1 and ITS-2 sequences, the Terranova larval type was resolved separately from Pseudoterranova spp. suggesting they do not belong to the genus Pseudoterranova.
As reviewed in the Introduction, Australian Terranova larval types could potentially be larval stages of Pseudoterranova spp., Terranova spp., or Pulchrascaris spp. Species associated with these genera have been listed in Table 3. Comparison of ITS sequence of Terranova larval type found in the present study with those of Pseudoterranova spp. available in GenBank (Table 4) shows a considerable nucleotide difference of 38.9–39.8% in both ITS-1 and ITS-2 regions. This is greater than nucleotide difference found for distinct species within a genus of family Anisakidae (Shamsi et al., 2009b) suggesting Terranova larval type in the present study does not belong to the genus Pseudoterranova.
| Terranova larval type in the present study | ||
|---|---|---|
| ITS-1 | ITS-2 | |
| P. azarsi | 39.8 | 46.7 |
| P. bulbosa | 38.9 | 48.4 |
| P. cattani | 39.3 | 47.2 |
| P. decipiens sensu tricto | 39.0 | 48.3 |
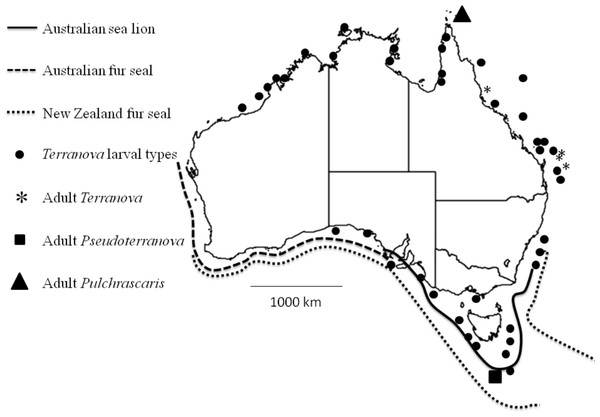
Figure 4: Map shows reported cases of Terranova larval types (circles), Adult Terranova spp (asterisk), adult Pseudoterranova spp (square), adult Pulchrascaris (triangle), distribution of Australian sea lion (solid line), Australian fur seal (square dots) and New Zealand fur seal (round dot).
To date, four species of Terranova have been reported from Australian sharks, T. galeocerdonis, T. ginglymostomae, T. pristis and T. scoliodontis (Bruce & Cannon, 1990). In addition, T. crocodyli was found in Australian crocodiles (Sprent, 1979). They all have a similar relationship between length of the intestinal caecum and ventriculus to the Terranova larval type in the present study. Pulchrascaris is a small genus in terms of number of species under family Anisakidae. Like members of the genus Terranova, Pulchrascaris spp. become adult in elasmobranches. There is intra/inter specific variation in the ratio of the intestinal caecum to ventriculus of Pulchrascaris spp. (Bruce & Cannon, 1990). Since there is no ITS sequences available for Terranova spp. or Pulchrascaris spp. in the GenBank database, the specific identity of the Terranova larval type found in the present study remains unknown and we are not able to associate these larvae to any Terranova spp. or Pulchrascaris spp. however, the present study, particularly the ITS sequence data, provides the essential information for future studies when the adult form is found and characterised.
This is the first report of a Terranova larval type from Abudefduf whitleyi, Carangoides fulvoguttatus, Caranx ignobilis, C. melampygus, Chaetodon flavirostris, Lutjanus argentimaculatus, L. bohar, Pristipomoides multidens, Scomber australasicus. Some of these fish, such as Australian mackerel (Scomber australasicus) are popular edible fish. Infection of those fish species that are not edible is also very important due to their role in the survival and transmission of Terranova larval type in the ecosystem.
Although the present study could not specifically identify the Terranova larval type in Australian waters, it could rule out the possibility of them being Pseudoterranova larvae which would have different implications for seafood and consumers’ safety and policy development in the country. It should be emphasized that it is very likely that Pseudoterranova larvae exist in Australian waters, infect some fish and await discovery. Their definitive hosts, Australian sea lion, Australian fur seal and New Zealand fur seal are found in southern coast of Australia (Fig. 4) and have been found to be infected with Pseudoterranova decipiens E (Bullini et al., 1997). However, given that in Australian waters the diversity of elasmobranch species is considerably higher (approximately 200 species, www.fishbase.net) than that of marine mammals (3 species of seals) our suggestion is that Terranova larval type in Australian waters is more likely to be a Terranova or a Pulchrascaris. To date there is no evidence that larval stage of Terranova or Pulchrascaris can cause infection in humans.