Molecular phylogeny and taxonomic review of Premnobiini Browne, 1962 (Coleoptera: Curculionidae: Scolytinae)
- Department of Entomology, Michigan State University, East Lansing, MI, USA
The taxonomy of Premnobiini is reviewed in the context of a molecular phylogeny including species of Ipini, Dryocoeotini, and Xyleborini. DNA data from COI, 16S, 28S, and CAD (~2640 characters) were generated for 79 species. Parsimony and Bayesian methods, using multiple sequence alignment methods and partitioning regimes, were used to reconstruct the phylogeny. The resulting topologies are generally congruent. Ipini is monophyletic along with all genera except Acathotomicus. Premnobiini is nested within Ipini and consists of two clades, which associate with the type species of Premnobius and Premnophilus, and with morphological diagnostic characters. The following taxonomic changes are made based on the recovered relationships. Premnophilus is resurrected as a valid genus and Premnobiini is considered a sub-tribe of Ipini.
Introduction
Premnobiini Browne (1962) consists of Premnobius Eichhoff (1879) and its 26 species that are endemic to sub-Saharan Africa; two of which are considered adventive to the Neotropics (Wood, 1982; Wood and Bright, 1992; Bright and Skidmore, 1997). Premnobius species are xylomycetophagus and are incestuous inbreeders, similar to Xyleborini (LeConte, 1876; Browne, 1961). This similarity in biology and gross morphology sparked debate on its taxonomic placement. Eichhoff (1878) originally suggested its affinity to Xyleborus Eichhoff (1864), however, subsequent researchers suggested alternative relationships with genera in the Corthylini, Dryocoeotini, and Xyloterini (Hagedorn, 1910; Hopkins, 1915). Schedl (1957) returned Premnobius to the Xyleborini and synonymized the genus with Xyleborus. Based on detailed examination of morphology and biology, Browne (1961) removed Premnobius from Xyleborus and described an additional closely related genus, Premnophilus Browne (1962). However, the tribal placement of Premnobius and Premnophilus remained uncertain despite the fact that Browne (1961) presented definitive morphological and behavioral evidence that these genera were unrelated to the Xyleborini. He suggested a new tribe, Premnobiini, for the genera and hypothesized its relationship between Ipini and Xyloterini. However, he postulated that these genera were Ipini, which converged to a xylomycetophagus habit (Browne, 1962). Premnophilus was soon synonymized with Premnobius because the morphological variation of its antennal club was considered within the range of variation observed for Xyleborus species (Schedl, 1964; Wood and Bright, 1992). Premnobiini was later formally established (Nobuchi, 1969), however, Wood (1982) did not recognize Premnobiini and placed Premnobius in Xyleborini stating that it represented the most primitive genus of the tribe. Phylogenetic studies including Premnobius cavipennis (Eichhoff, 1878) and various amount of molecular and morphological data demonstrated monophyly of Premnobius with Ipini genera (Normark et al., 1999; Farrell et al., 2001). Although no nomenclatural changes were made in these studies, subsequent researchers (Beaver, 2005; Bright and Torres, 2006; Alonso-Zarazaga and Lyal, 2009) recognized Premnobiini based on data from previous studies (Browne, 1961, 1962; Nobuchi, 1969; Normark et al., 1999). The most comprehensive phylogenetic analysis of scolytines placed Premnobius (two species) sister to Acanthotomicus and monophyletic with ten other Ipini species thus rendering Ipini paraphyletic (Jordal et al., 2011; Jordal and Cognato, 2012). The Premnobiini + Ipini clade was sister to the Xyleborini + Dryocoetini clade which both had 100% posterior probabilities and demonstrated, once again, the distant relation between Premnobiini and Xyleborini (Jordal and Cognato, 2012).
The previous phylogenetic studies did not address taxonomic or nomenclatural issues suggested by the resulting phylogenies (Normark et al., 1999; Farrell et al., 2001; Jordal et al., 2011; Jordal and Cognato, 2012). This study provides a more detailed phylogenetic analysis designed to address the tribal and generic status of Premnobiini and Premnophilus, respectively. Phylogenies reconstructed with molecular data for 79 species representing Dryocoetini (2 spp.), Premnobiini (15 spp.), Ipini (59 spp. including all genera), and Xyleborini (3 spp.) support the monophyly of Premnophilus and the inclusion of Premnobiini within Ipini. Taxonomic and nomenclatural changes concerning the Premnobiini are made based on these results.
Materials and Methods
Taxa and DNA Sequencing
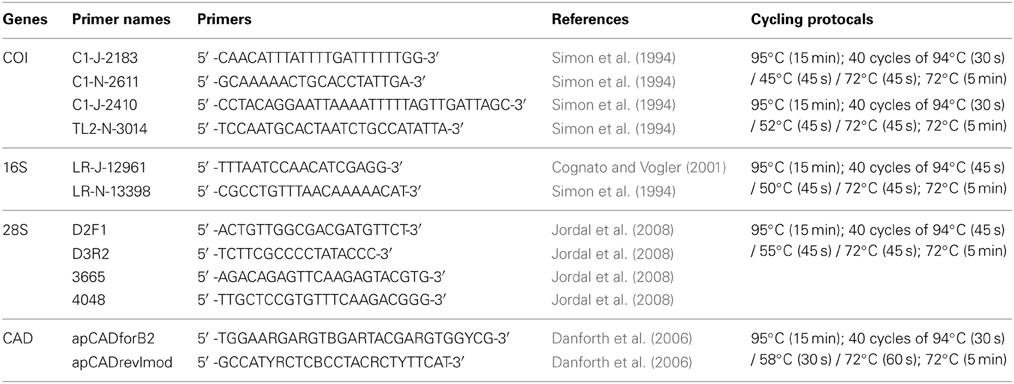
DNA data for mtDNA cytochrome oxidase I (COI), mtDNA ribosomal 16S, nuclear ribosomal 28S (D2 and D3 regions), and nuclear protein coding gene CAD were generated for 22 species of Premnobiini and Acanthotomicus. The inclusion of existing data (Cognato and Sun, 2007; Jordal and Cognato, 2012) and the generation of CAD data created a data set of 79 species, which represented all Ipini genera and outgroup Dryocoetini and Xyleborini species (Table 1). Beetle specimens were collected over 20 years from various countries by AIC and his collaborators (Table 1). DNA was extracted from frozen and pinned specimens following the Qiagen (Hilden, Germany) tissue extraction protocol using Qiagen blood and tissue kits. Tissue was digested from the head and pronotum. These parts were recovered at the end of the extraction procedure. The head and pronotum and the remaining body were pinned and vouchered at the A.J. Cook Arthropod Research Collection, Michigan State University. Using the purified DNA, partial gene regions of COI, 16S, 28S, CAD were amplified using the primers and protocol listed in Table 2. Unincorporated nucleotides, primers and taq remaining in the PCR reactions were neutralized with ExoSAP-IT (USB Corp., Cleveland, OH) following the manufacturer protocols. Both strands of the clean PCR products were directly sequenced using BigDye Terminator v.1.1 (Applied Biosystems, Foster City, CA, USA) cycle sequencing kit and visualized on an ABI 3730 (Applied Biosystems) at the Research Technology Support Facility at Michigan State University. The DNA sequences were compiled and edited with Sequencher (GeneCodes, Ann Arbor, MI, USA) and archived at Genbank (Table 1).
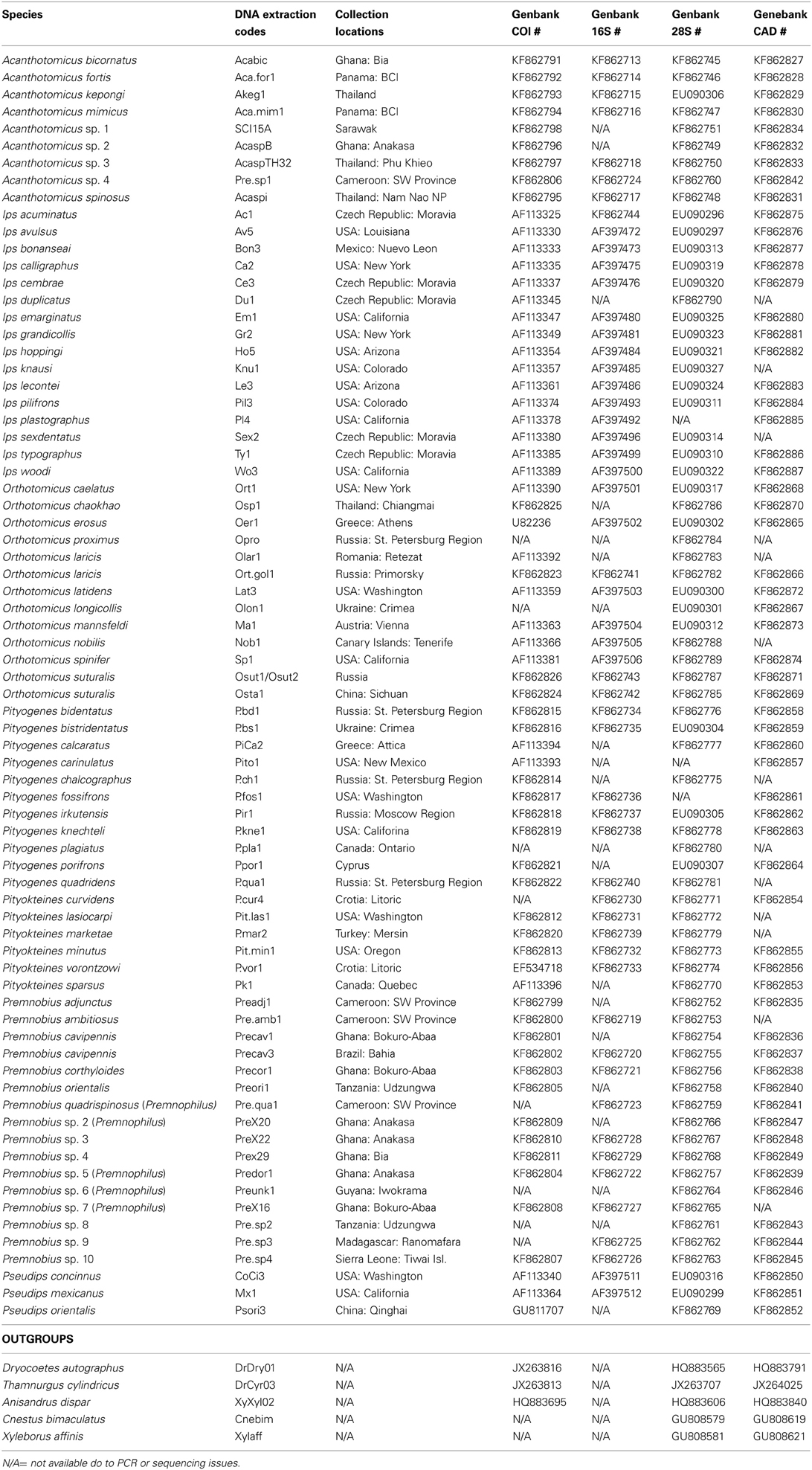
Table 1. Specimens included in current analyses with associated geographic localities and Genbank accession numbers.
DNA Sequence Alignment and Phylogenetic Analyses
Protein coding genes were aligned without the aid of software computation given that insertion/deletions and introns were not observed. Ribosomal DNA sequences varied in their length. These sequences were manually aligned based on previous published alignments (Cognato and Vogler, 2001; Jordal et al., 2008) and using the alignment software, MUSCLE, with default parameters (Edgar, 2004). These data sets were treated separately in subsequent phylogenetic analyses.
Parsimony and maximum likelihood optimality criteria were used to reconstruct phylogenies. Parsimony analyses using PAUP* 4.0 b10 PPC (Swofford, 2002) consisted a heuristic search with 500 stepwise random addition replicates keeping only 500 suboptimal trees per replicate and the default settings with the manually and MUSCLE aligned data sets. These analyses were repeated while treating gap positions as 5th characters states. Bootstrap values were calculated with 500 pseudoreplicates for the four data treatments. Partition Bremer support was calculated with TreeRot v.2 (Sorenson, 1999). For the manually and MUSCLE aligned data sets, maximum likelihoods were estimated via Bayesian analysis using Mr. Bayes 3.2.2 (Ronquist et al., 2012). These analyses consisted of two simultaneous runs of four Metropolis-Coupled Markov chain Monte Carlo searches (one cold, three heated). Each search comprised 10 million generations which was sampled every 100th iteration. The data were partitioned by codon positions and ribosomal genes and each partition was allowed to independently evolve under a general time reversible (GTR + I + Γ) model. All parameters reached stability within 10 million generations and the split distribution between runs did not vary much (mean standard deviation between runs = 0.002). Bayesian posterior probabilities of clades were calculated by a majority-rule consensus of those trees after the burn-in (for both runs 75000 trees).
Results and Discussion
Phylogenetic Analyses
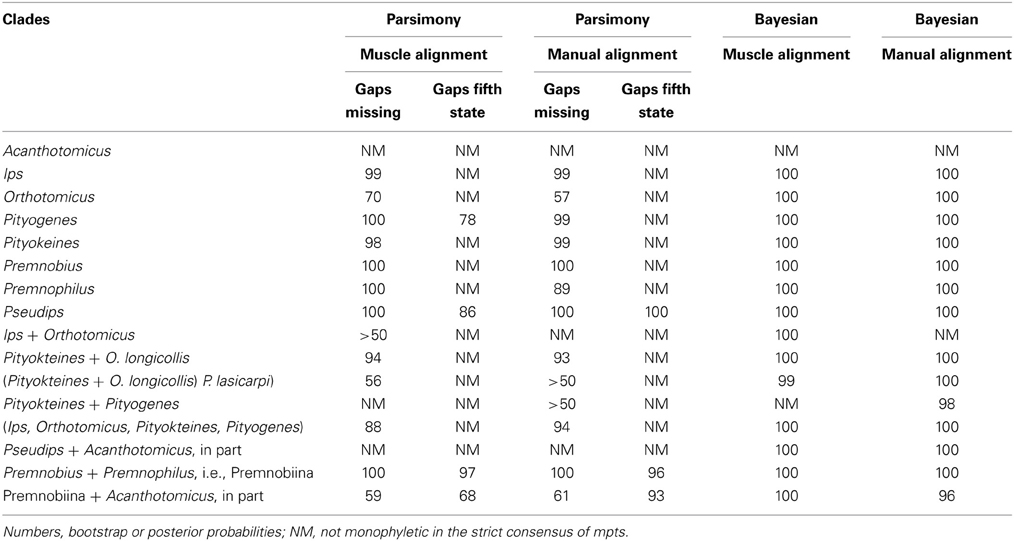
The combination of analyses and alignments resulted in tree topologies that differed by various sub-clades but were generally similar (e.g., Figures 1–3). For example, Ipini including Premnobiini was monophyletic and the clade consisting of Ips, Orthotomicus, Pityogenes, and Pityokteines was found in the majority of analyses (Table 3). Treating gapped positions as 5th character states in the parsimony analyses resulted in trees with many non-monophyletic genera. The monophyly of these genera is not suspect given the many morphological diagnostic and synapomorphic characters recognized for each genus (e.g., Hopping, 1963; Wood, 1986; Cognato, 2000). Given the lack of taxonomic congruence, the analyses that treated gapped positions as 5th character states were not given further consideration.
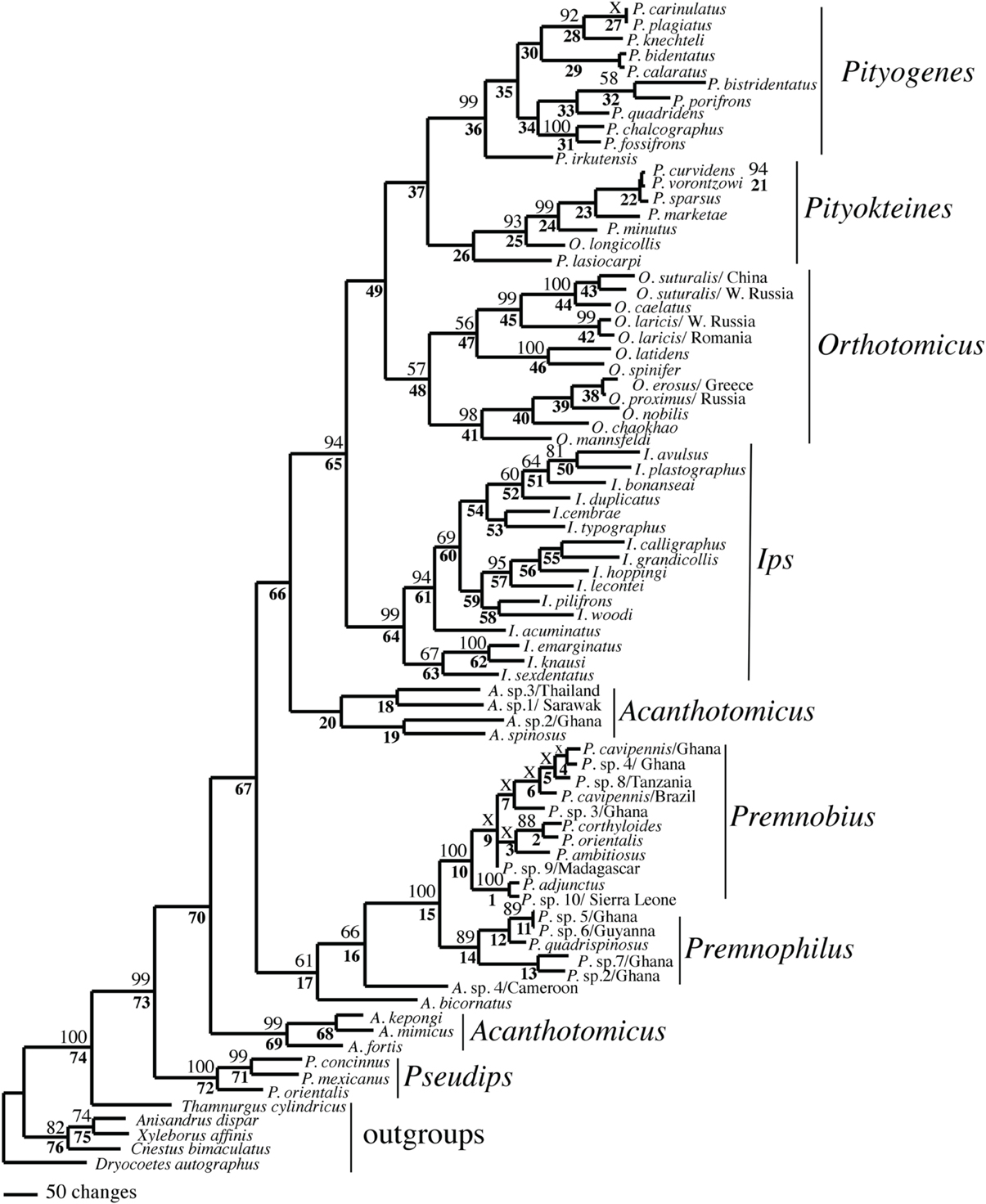
Figure 1. Phylogeny of Ipini and outgroup species; 1 of 87 most parsimonious trees based on the manually aligned data. Clades marked by “X” were not resolved in the strict consensus of the 87 mpts. Numbered clades are referenced in Table 4. Numbers = bootstrap values and clades without numbers had > 50% bootstrap values.
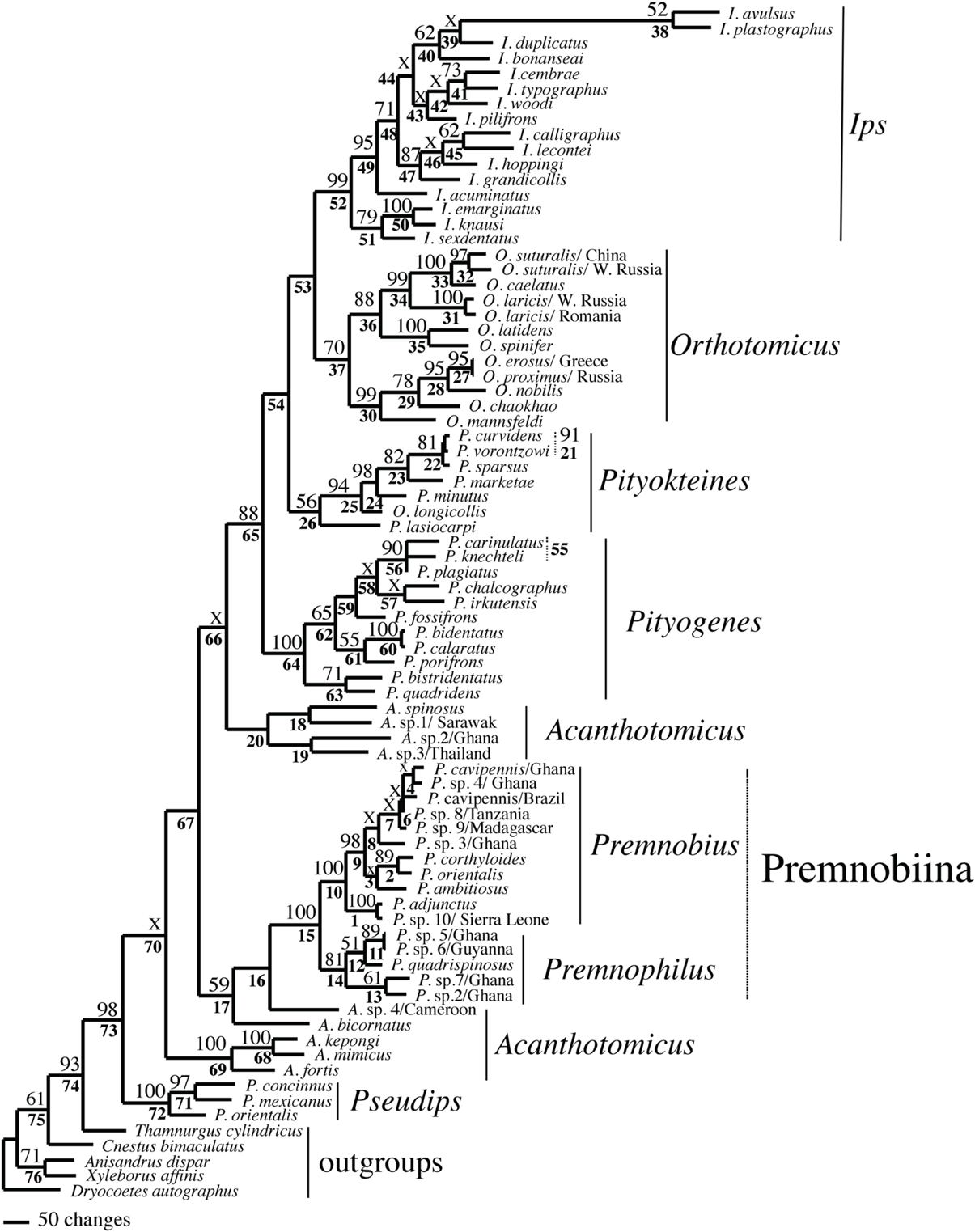
Figure 2. Phylogeny of Ipini and outgroup species; 1 of 756 most parsimonious trees based on the MUSCLE aligned data. Clades marked by “X” were not resolved in the strict consensus of the 756 mpts. Numbered clades are referenced in Table 5. Numbers = bootstrap values and clades without numbers had >50% bootstrap values.
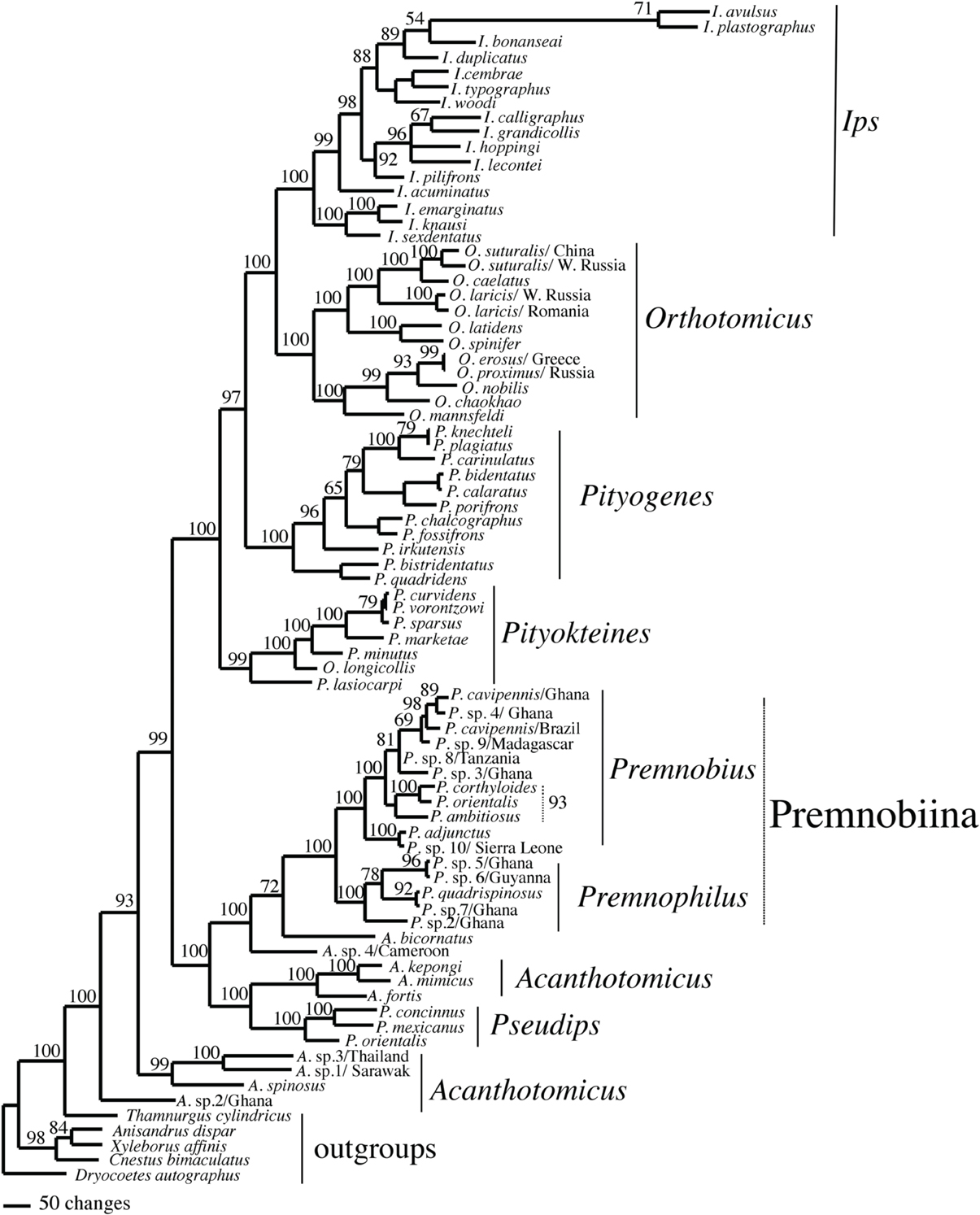
Figure 3. Majority rule consensus of 75,000 trees found in a Bayesian analysis of Ipini and outgroup species. Numbers are posterior probabilities.
The alignments affected the distribution of branch support among the data partitions. For the manual alignment 16S exhibited two-three times more support than the other data sets (Table 4). Data interaction changed for the MUSCLE alignment. CAD and 28S exhibited two-six times more support than COI and 16S and support from 16S decreased by a third (Table 5). Also the MUSCLE alignment supported three times more clades as compared to the manual alignment (Tables 4 and 5). Changes in support given different alignments have been observed in other studies (e.g., Cognato and Vogler, 2001; Damgaard et al., 2004). An increase in support values was interpreted as increased concordance of data interaction (Cognato and Vogler, 2001). Thus, the parsimony and Bayesian phylogenies (Figures 2, 3) based on the MUSCLE aligned data with gap positions scored as missing were considered as better hypotheses as compared to the trees found with the manually aligned data.
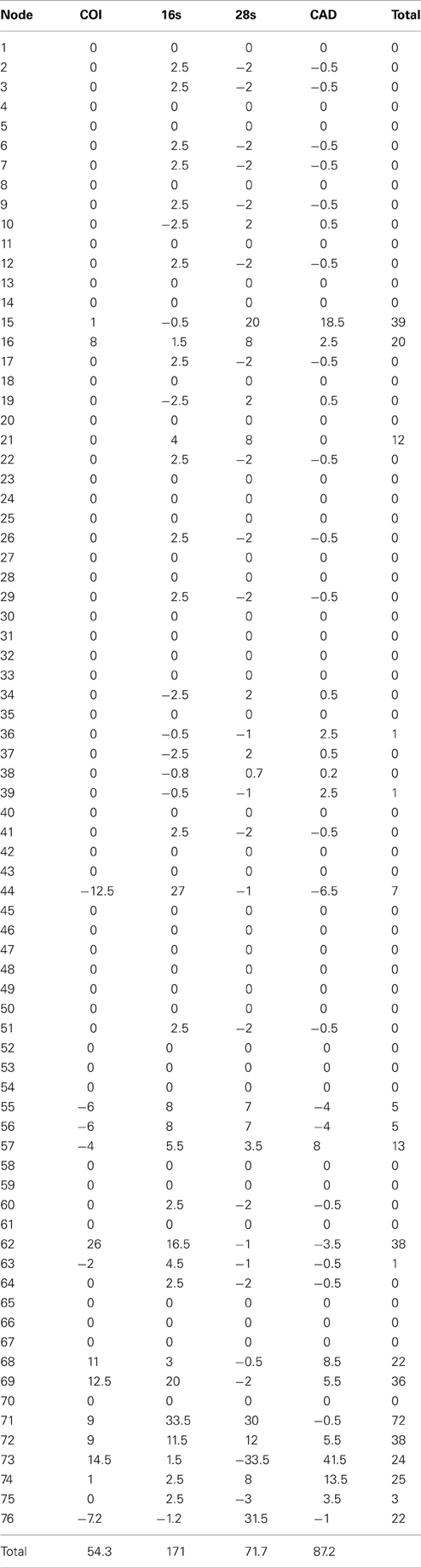
Table 4. Partition bremer support for 1 of 87 most parsimonius trees reconstructed with the manually aligned data (Figure 1).
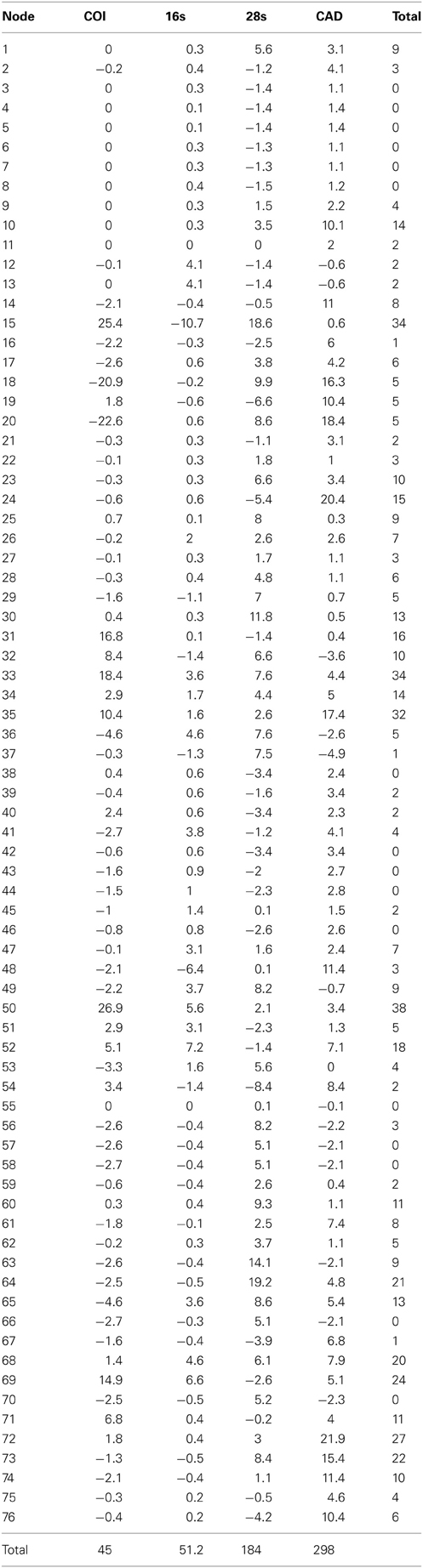
Table 5. Partition bremer support for 1 of 756 most parsimonius trees reconstructed with the MUSCLE aligned data (Figure 2).
Both analyses mostly recovered consistent results with all genera (except Acanthotomicus) and Premnobiina as monophyletic. However, the relationships of these genera differed between the analyses. Acanthotomicus, in part, or Pseudips was sister to the other Ipini genera, however, there was no to little support for this position of the latter genus (Figure 1). The position of Pseudips imbedded within Acathotomicus agreed with previous phylogenies that demonstrated that Pseudips was not monophyletic with Ips (Cognato, 2000) or other Holarctic Ipini genera (Jordal and Cognato, 2012). This result also supported the validity of Pseudips as distinct from Orthotomicus (Wood, 2007; Alonso-Zarazaga and Lyal, 2009; Knížek, 2011).
Acanthotomicus was polyphyletic (Figures 1–3) and confirmed similar results of a study which included fewer Acanthotomicus specimens (Jordal and Cognato, 2012). Clades did not associate with geographic region because several clades comprised a mixture of species from Asia, Africa and/or South America (Figures 1–3). This was not expected because the pantropical distribution of the genus predicts that genetic isolation would associate with geographic distance (Avise, 2000). Although there is a gestalt to their appearance which suggests monophyly of the genus, groups of species differ in details of antennal and elytral morphology. For example, a pattern of morphological concordance was present, for a clade including A. spinosus, A. sp. 1/Sarawak, and A. sp. 3/ Thailand (Figure 3) which have long spines on their elytral declivity. Additional patterns may emerge with the inclusion of the ~90% remaining Acanthotomicus species in future phylogenetic analyses which will provide better hypotheses of interspecific and intergeneric relationships of Acanthotomicus. However, it appears unlikely that a monophyletic Acanthotomicus will be resolved, given the high support for these phylogenies (Figure 3) and others (Jordal and Cognato, 2012) thus a thorough revision of the genus will be necessary.
The remaining Holarctic genera, Ips, Orthotomicus, Pityogenes, and Pityokteines, were monophyletic, except for one Orthotomicus species included in Pityokteines, but the relationships among the genera varied and were not well-supported (Table 3, Figures 1, 2). Details of the species relationships for each genus will be the subject of future studies. However, two conspicuous results will be discussed. First, O. longicollis (Gyllenhal, 1827) was monophyletic with Pityokteines (Figures 1–3) as previously observed within a 28S based phylogeny (Jordal et al., 2008). This species has an obliquely truncated antennal club with inconspicuous sutures similar to the antennal club of Pityokteines. The generic placement of O. longicollis in Pityokteines appears appropriate but additional nucleotide data is needed to confirm this relationship. Second, Pityokteines lasiocarpi (Swaine, 1916), which was once considered a monotypic genus, Orthotomides Wood (1951), was sister to the Pityokteines species including O. longicollis (Figures 1–3). Although the structure of the antennae of P. lasisocarpi is not typical of Pityokteines, there is currently little reason for the resurrection of Orthotomides.
The clade including Premnobius and Premnophilus was well-supported (Table 3, Figures 1–3). This clade was sister to either of two African Acanthotomicus species and well-imbedded amongst other Ipini genera. Given this phylogenetic position, the tribal status of this clade is unjustified; see below. Premnophilus was reciprocally monophyletic with Premnobius and included P. quadrispinosus (Schedl, 1938) and three unidentified species (likely undescribed). Given a well-supported clade and morphological diagnostic characters, Premnophilus is recognized as distinct from Premnobius; see below. Premnobius was also well-supported (Figures 1–3) and include species that were characterized by a carinate lateral eytral margin. These species resembled P. cavipennis and differed by minor morphological differences of the elytral declivity (clade 9, Figure 2). One species rendered P. cavipennis paraphyletic, which suggests the need of a revision. Terminal branch lengths of unidentified species were similar in length to terminal branch lengths of known species (e.g., P. sp. 3/Ghana and P. ambitiosus) (Figure 3). If branch lengths along with minor morphological differences are taken as a proxy for species delimitation, then a greater Premnobius species diversity awaits discovery.
Taxonomy
Premnophilus Browne, genus bona (Figures 4, 5)
Premnophilus (Browne, 1962): 79. Type species: Xyleborus joveri (Schedl, 1951)= Premnobius quadrispinosus (Schedl, 1938); original designation.

Figure 4. Lateral view of the holotype of Premnophilus quadrispinosus, the type species of Premnophilus. Specimen length is approximately 1.5 mm.
Diagnosis
Browne (1962) thoroughly described and illustrated the genus. To emphasize, Premnophilus differs from Premnobius by the obliquely truncated antennal club without obvious sutures. The anterior edge of the first corneous segment is sinuous. In Premnobius the antennal club is flat with two procurved sutures and the anterior edge of the first corneous segment is procurved. Also, the lateral edge of the eytral declivity for Premnophilus is rounded as compared to the acute lateral edge of the eytral declivity for Premnobius.
Included species
I have observed specimens of the listed species. Two or three likely undescribed species (e.g., P. sp. 5/Ghana, P. sp. 6/Guyana, Figure 1) also belong to Premnophilus. A revision of this genus will occur in a subsequent publication (Cognato, in prep.). Complete references are given in Wood and Bright (1992).
Premnophilus quadrispinosus (Schedl), new combination
Premnobius quadrispinosus Schedl, 1938: 461.
Xyleborus joveri Schedl, 1951: 41–42.
Premnophilus perspinidens (Schedl), new combination
Xyleborus perspinidens Schedl, 1957: 107.
Premnobiina, status novo
Included genera.
Premnobius Eichhoff, 1879.
Premnophilus Browne, 1962.
As indicated in the results, Premnobius and Premnophilus are monophyletic and sister to Acanthotomicus, in part (Figures 1–3). Consideration of these genera as a tribe renders Ipini paraphyletic. To remedy this issue, Premnobiini is recognized as a sub-tribe of Ipini. Alternatively, clades of Acanthotomicus and Pseudips could be recognized as tribes to preserve the monophyly of Ipini and the tribal status of Premnobiini. However, this action is not advisable because of the limited sample of Acanthotomicus species included in these analyses.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I thank Sarah M. Smith, Bjarte H. Jordal, and the Review Editors for helpful discussion of this topic and for review of previous manuscripts. I appreciate the collecting efforts of Jiri Hulcr, Bjarte H. Jordal, and Sarah M. Smith, which helped make this study possible. This study was supported in part by the Norwegian Research Council (214232) and the University of Bergen, Norway (F20) awarded to Bjarte H. Jordal. and NSF-PEET (DEB 0328920) awarded to Anthony I. Cognato.
References
Alonso-Zarazaga, M., and Lyal, C. H. C. (2009). A catalogue of family and genus group names in scolytinae and platypodinae with nomenclatural remarks (Coleoptera: Curculionidae). Zootaxa, 2258, 1–134.
Avise, J. C. (2000). Phylogeography: The History and Formation of Species. Cambridge, MA: Harvard University Press.
Beaver, R. A. (2005). A remarkable new species of Cyclorhipidion Hagedorn, and new records of bark and ambrosia beetles from Gabon (Coleoptera: Curculionidae, Scolytinae and Platypodinae). Entomol. Mon. Mag. 141, 113–119.
Bright, D. E., and Skidmore, R. E. (1997). A Catalog of Scolytidae and Platypodidae (Coleoptera), Supplement 1 (1990–1994). Ottawa, ON: NRC Research Press, 1–368.
Bright, D. E., and Torres, J. A. (2006). Studies on West Indian Scolytidae (Coleoptera) 4. A review of the Scolytidae of Puerto Rico, U.S.A. with descriptions of one new genus, fourteen new species and notes on new synonymy (Coleoptera: Scolytidae). Koleopterologische Rundschau 76, 389–428.
Browne, F. G. (1961). The generic characters, habits and taxonomic status of Premnobius Eichhoff (Coleoptera, Scolytidae). Fourth WATBRU Rep. 1961, 45–51.
Cognato, A. I. (2000). Phylogenetic analysis reveals new genus of Ipini bark beetle (Scolytidae). Ann. Entomol. Soc. Am. 93, 362–366. doi: 10.1603/0013-8746(2000)093[0362:PARNGO]2.0.CO;2
Cognato, A. I., and Sun, J. H. (2007). DNA based cladograms augment the discovery of a new Ips species from China (Coleoptera: Curculionidae:Scolytinae). Cladistics 23, 539–551. doi: 10.1111/j.1096-0031.2007.00159.x
Cognato, A. I., and Vogler, A. P. (2001). Exploring data interaction and nucleotide alignment in a multiple gene analysis of Ips (Coleoptera: Scolytinae). Syst. Biol. 50, 758–780. doi: 10.1080/106351501753462803
Damgaard, J., Andersen, N., and Meier, M. R. (2004). Combining molecular and morphological analyses of water strider phylogeny (Hemiptera-Heteroptera, Gerromorpha): effects of alignment and taxon sampling. Syst. Entomol. 30, 289–309 doi: 10.1111/j.1365-3113.2004.00275.x
Danforth, B. N., Fang, J., and Sipes, S. (2006). Analysis of family-level relationships in bees (Hymenoptera: Apiformes) using 28S and two previously unexplored nuclear genes: CAD and RNA polymerase II. Mol. Phylogenet. Evol. 39, 358–372. doi: 10.1016/j.ympev.2005.09.022
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Eichhoff, W. J. (1864). Uber die Mundtheile und die Fuhlerbildung der europaischen Xylophagi sens strict. Berliner Entomol. Zeits. 8, 17–46. doi: 10.1002/mmnd.18640080103
Eichhoff, W. J. (1878). Ratio, descriptio, emendatio eorum Tomicinorum qui sunt in Dr. medic. Chapuisi et autoris ipsius collectionilbus et quos praeterea recognovit. Mémoires de la Société Royale des Sciences de Liège, 81, 1–531.
Eichhoff, W. J. (1879). Zur Entwicklungsgeschichte der Borkenkafer. Stettiner Entomogische Zeitung 40, 501–506.
Farrell, B. D., Sequeira, A. S., O'Meara, B. C., Normark, B. B., Chung, J. H., and Jordal, B. H. (2001). The evolution of agriculture in beetles (Curculionidae: Scolytinae and Platypodinae). Evolution 55, 2011–2027. doi: 10.1111/j.0014-3820.2001.tb01318.x
Gyllenhal, L. (1827). Insecta Svecica descripta a Leonardo Gyllenhal. Clasis I Coleoptera sive Eleuterata. Tomi 1, Pars IV. Lipsiae: Friedericum Fleischer. 761.
Hagedorn, J. M. (1910). “Ipidae. Pars 4,” in Coleopterorum Catalogus, ed S. Schenkling (Berlin: W. Junk), 134.
Hopkins, A. D. (1915). Contributions toward a monograph of the scolytid beetles, Part II Preliminary classification of the superfamily Scolytoidea. US Depart. Agric. Tech. Bull. 17, 165–232.
Hopping, G. R. (1963). Generic characters in the tribe Ipini (Coleoptera: Scolytidae), with a new species, a new combination, and new synonymy. Can. Entomol. 95, 61–68. doi: 10.4039/Ent9561-1
Jordal, B., Gillespie, J. J., and Cognato, A. I. (2008). Secondary structure alignment and direct optimization of 28SrDNA sequences provide limited phylogenetic resolution in bark and ambrosia beetles (Curculionidae: Scolytinae). Zool. Scr. 37, 43–56. doi: 10.1111/j.1463-6409.2007.00306.x
Jordal, B. H., and Cognato, A. I. (2012). Molecular phylogeny of bark beetles reveals multiple origins of fungus farming during periods of global warming. BMC Evol. 12:133. doi: 10.1186/1471-2148-12-133
Jordal, B. H., Sequeira, A., and Cognato, A. I. (2011). The age and phylogeny of wood boring weevils and the origin of subsociality. Mol. Phylogenet. Evol. 59, 708–724. doi: 10.1016/j.ympev.2011.03.016
Knížek, M. (2011). “Scolytinae,” in Catalogue of Palaearctic Coleoptera, Vol. 7, eds I. Löbl and A. Smetana (Stenstrup: Apollo Books), 204–250.
LeConte, J. L. (1876). “Family IX. Scolytidae,” in The Rhynchophora of America North of Mexico. eds J. L. LeConte and G. H. Horn, (Philadelphia, PA: American Philosophical Society), 341–391, Appendix: p. 426.
Nobuchi, A. (1969). A comparative morphological study of the proventriculus in the adult of the superfamily Scolytoidea (Coleoptera). Gov. For. Exp. Station Bull. (Tokyo) 224, 39–110.
Normark, B. B., Jordal, B. H., and Farrell, B. D. (1999). Origin of a haplodiploid beetle lineage. Proc. R. Soc. B 266, 2253–2259. doi: 10.1098/rspb.1999.0916
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Schedl, K. E. (1957). Scolytoidea nouveaux du Congo Belge, II Mission R Mayne – K E Schedl 1952. Ann. Mus. Congo Belge Tervuren (Belgique) Sci. Zool. 56, 1–1162.
Schedl, K. E. (1964). West African bark and timber beetles, I. Reichenbachia Mus. Tierkd. Dresden 4, 39–52.
Simon, C., Frati, F., Beckenbach, A., Crespi, B., Liu, H., and Flook, P. (1994). Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 87, 651–701.
Swaine, J. M. (1916). New species of the family Ipidae, pt. III. Can. Entomol. 48, 181–192. doi: 10.4039/Ent48181-6
Swofford, D. (2002). PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods) Version 4. Sunderland, MA: Sinauer Associates.
Wood, S. L. (1951). Two new species and a new genus of Scolytidae (Coleoptera) from Utah. Kansas Entomol. Soc. J. 24, 31–32.
Wood, S. L. (1982). The bark and ambrosia beetles of North America (Coleoptera: Scolytidae), A Taxonomic Monograph. Great Basin Nat. Mem. 6, 1–1359.
Wood, S. L. (1986). A reclassification of the genera of Scolytidae (Coleoptera). Great Basin Nat. Mem. 10, 1–126.
Keywords: Ipini, systematics, taxonomy, nomenclature, COI, 16S, 28S, CAD
Citation: Cognato AI (2013) Molecular phylogeny and taxonomic review of Premnobiini Browne, 1962 (Coleoptera: Curculionidae: Scolytinae). Front. Ecol. Evol. 1:1. doi: 10.3389/fevo.2013.00001
Received: 29 October 2013; Paper pending published: 04 November 2013;
Accepted: 17 November 2013; Published online: 29 November 2013.
Edited by:
Alejandro P. Rooney, U.S. Department of Agriculture, USACopyright © 2013 Cognato. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anthony I. Cognato, Department of Entomology, Michigan State University, 288 Farm Lane Road, East Lansing, MI 48824, USA e-mail: cognato@msu.edu
 Anthony I. Cognato
Anthony I. Cognato