Chloroplast Genomes of the Green-Tide Forming Alga Ulva compressa: Comparative Chloroplast Genomics in the Genus Ulva (Ulvophyceae, Chlorophyta)
- 1CAS Key Laboratory of Marine Ecology and Environmental Sciences, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 2Marine Ecology and Environmental Science Laboratory, Pilot National Laboratory for Marine Science and Technology (Qingdao), Qingdao, China
- 3Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao, China
- 4Spelman College, Atlanta, GA, United States
To understand the evolution of Ulva chloroplast genomes at intraspecific and interspecific levels, in this study, three complete chloroplast genomes of Ulva compressa Linnaeus were sequenced and compared with the available Ulva cpDNA data. Our comparative analyses unveiled many noticeable findings. First, genome size variations of Ulva cpDNAs at intraspecific and interspecific levels were mainly caused by differences in gain or loss of group I/II introns, integration of foreign DNA fragments, and content of non-coding intergenic spacer regions. Second, chloroplast genomes of U. compressa shared the same 100 conserved genes as other Ulva cpDNA, whereas Ulva flexuosa appears to be the only Ulva species with the minD gene retained in its cpDNA. Third, five types of group I introns, most of which carry a LAGLIDADG or GIY-YIG homing endonuclease, and three of group II introns, usually encoding a reverse transcriptase/maturase, were detected at 26 insertion sites of 14 host genes in the 23 Ulva chloroplast genomes, and many intron insertion-sites have been found for the first time in Chlorophyta. Fourth, one degenerate group II intron previously ignored has been detected in the infA genes of all Ulva species, but not in the closest neighbor, Pseudoneochloris marina, and the other chlorophycean taxa, indicating that it should be the result of an independent invasion event that occurred in a common ancestor of Ulva species. Finally, the seven U. compressa cpDNAs represented a novel gene order which was different from that of other Ulva cpDNAs. The structure of Ulva chloroplast genomes is not conserved, but remarkably plastic, due to multiple rearrangement events.
Introduction
The green algal class Ulvophyceae, as one of the five classes of green algae in the core Chlorophyta (Cocquyt et al., 2010; Lang and Nedelcu, 2012; Fučíková et al., 2014), encompasses at least ten orders and more than 1,900 species (Mine et al., 2008; Leliaert et al., 2012; Guiry and Guiry, 2021). To date, the chloroplast genomes (cpDNAs) in Ulvophyceae have been sequenced for at least 65 taxa including 37 in Bryopsidales, 15 in Ulvales (14 Ulva species and one Pseudoneochloris), seven in Ulotrichales, two in Ignatiales, two in Oltmannsiellopsidales, one in Trentepohliales and one in Cladophorales. The available complete cpDNAs in Ulvophyceae display typical circular chloroplast genomes, with the exception of Boodlea composita (Cladophorales) cpDNA which was fragmented into multiple hairpin chromosomes and had a highly reduced gene repertoire (Del Cortona et al., 2017). The circular ulvophycean chloroplast genomes range in size from the smallest one with 74.5 kb in Callipsygma wilsonis (Bryopsidales) (Cremen et al., 2018) to the largest with 399.4 kb in Trentepohlia odorata (Trentepohliales) (Zhu et al., 2019). These ulvophycean chloroplast genomes display great variations at the interspecific level in gene content, gene density, content of group I and II intron, gene order, copies of large inverted repeat (IR), and genome architecture (Turmel et al., 2017; Turmel and Lemieux, 2018; Kim et al., 2019).
The macroalgal genus Ulva Linnaeus is one of the most speciose genera in the Ulvophyceae, and contains more than 80 species currently accepted taxonomically, ranking the fourth only next to Cladophora (197 species), Codium (144 species) and Caulerpa (104 species) (Guiry and Guiry, 2021). Ulva species have a wide geographic distribution in marine and estuarine environments all over the world, and share similar reproductive strategies and life cycle (Balar and Mantri, 2020). Their morphologically simple thalli are composed of only three differentiated cell types (Spoerner et al., 2012). Ulva species could serve as a group of useful model organisms to study the algal development, morphogenesis, and the interaction between Ulva and their symbiotic bacteria (Wichard et al., 2015). Ulva species usually exhibit great morphological plasticity due to abiotic factors such as temperature, salinity, irradiance, wave exposure, growth phase, and nutrient content (Blomster et al., 2002; Gao et al., 2016) and biological factors such as symbiotic bacteria and grazing (Marshall et al., 2006). Morphological, anatomical, cytological, mating, and molecular characteristics have been used to evaluate species concepts in this genus (Liu et al., 2013; Hiraoka et al., 2017; Hughey et al., 2019), while molecular data has become a more reliable and commonly used method for species identifications.
Ulva compressa Linnaeus is a common marine green macroalga distributed in coasts of Asia, Europe, and America (Guiry and Guiry, 2021). U. compressa displays high intraspecific morphological plasticity from the filamentous, highly branched morphotype, to the ribbon, blade-like morphotype, to the foliated free-floating morphotype (Blomster et al., 1998; Steinhagen et al., 2019a, b; Liu F. et al., 2020). In eutrophic water body, this alga could grow rapidly and accumulate massive biomass, causing notorious green tides (Blomster et al., 2002). Recently, Ulva mutabilis Föyn has been regarded as a taxonomic synonym of U. compressa Linnaeus based on mating test and molecular phylogenetics (Steinhagen et al., 2019a). This alga is the first green seaweed to have its nuclear genome sequenced, and its haploid genome is 98.5 Mbp in size and harbors 12,924 protein-coding genes (PCGs) (De Clerck et al., 2018).
Previously, U. compressa samples with three different morphotypes were collected from China and the United States, and their mitochondrial genomes were sequenced and compared at intraspecific and interspecific levels. The U. compressa mitogenomes displayed substantial variation in genome size, gene content, and intron content, due to different acquisitions of foreign DNA fragments, gain or loss of group I/II introns, and non-coding intergenic spacer regions (Liu F. et al., 2020). To understand the evolution of Ulva chloroplast genomes at intraspecific and interspecific levels, in this study, we assembled and annotated the complete chloroplast genomes of three U. compressa samples from China and the United States, and performed a comparative analysis with the available Ulva cpDNA data deposited in GenBank.
Materials and Methods
Sampling and Species Identification
Algal thallus of Ulva compressa Linnaeus with foliated, distromatic blade (Uco1) was sampled from Swansboro (34°41′N, 77°06′W), NC, United States in Apr. 2015. The sample was preserved in silica gel and as herbarium vouchers, was submitted to the University of Alabama Herbarium (UNA00072687). The genomic DNA was extracted using a Qiagen Plant DNA Extraction Kit (QIAGEN, Valencia, CA, United States). The U. compressa thallus with ribbon, distromatic blade (Uco2) was collected in Huiquan Bay (36°03′N, 120°20′E), Qingdao, Shandong, China in Apr. 2017. The U. compressa thallus with filamentous, tubular blade and dense branches (Uco3) was sampled in Subei Shoal (33°30′N, 120°57′E), Jiangsu, China in May 2016. The algal samples were kept in coolers (5-8°C) and transported to laboratory in IOCAS within 48 h after collection. Fresh algal tissue from one individual thallus was used for DNA extraction using a Plant Genomic DNA Kit (Tiangen Biotech, Beijing, China).
Species identification of three U. compressa morphotypes was conducted based on phylogenetic analyses of two common DNA markers including the internal transcribed spacer DNA (ITS) region including the 5.8S rDNA gene, and the plastid-encoded large subunit of the ribulose 1,5-bisphosphate carboxylase gene (rbcL) (Hayden and Waaland, 2002). Primers sequences and polymerase chain reaction (PCR) amplification were used according to our previous study (Liu et al., 2010). Sequence datasets of our samples and other data from GenBank were aligned using MEGA 7.0 (Kumar et al., 2016). The maximum likelihood (ML) tree was constructed with 1,000 bootstrap replicates based on the Kimura two-parameter model (Kimura, 1980). The identification results confirmed that these three samples were U. compressa (Liu F. et al., 2020).
DNA Sequencing and Assembly
The concentration and quality of isolated DNA were evaluated with a NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States). For the sample from the United States, paired end reads (150 bp) were sequenced at Cold Spring Harbor Laboratory on an Illumina MiSeq platform. For the samples from China, the purified DNA was fragmented into 350 bp and used to construct short-insert libraries. The short fragments were sequenced using an Illumina Hiseq 4000 sequencing platform. Poor quality sequences and sequencing adapters were removed using Trim Galore! v0.3.71. The U. compressa chloroplast genomes were constructed using a combination of de novo and reference-guided assemblies. For the sample from the United States, genome assembly was done with both A5 (Tritt et al., 2012) and Geneious R7 (Kearse et al., 2012). For two samples from China, the filtered reads were assembled into contigs using SOAPdenovo2.04 (Luo et al., 2012). Incomplete genomes were closed by iteratively mapping the trimmed reads on to the contig with Geneious 7.1.
Annotation of Ulva Chloroplast Genomes
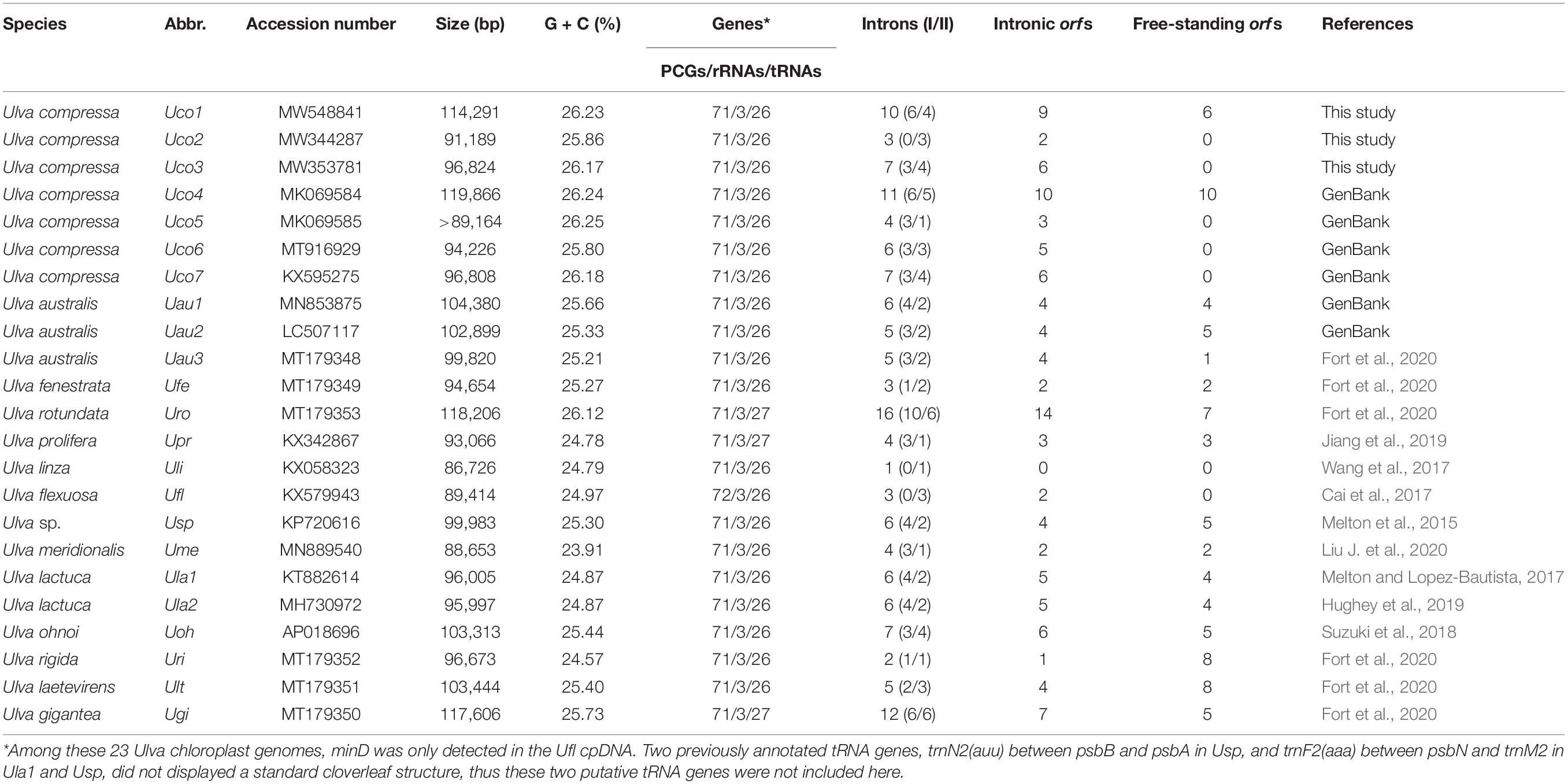
Protein-coding genes (PCGs) were annotated by Open Reading Frame (ORF) Finder at the National Center for Biotechnology Information (NCBI), DOGMA (Wyman et al., 2004) and ORF finder in Geneious 7.1. Ribosomal RNA (rRNA) genes were identified by a BLAST of the non-redundant databases at the NCBI (Altschul et al., 1997) and by comparing newly sequenced U. compressa cpDNAs with rRNA genes from other Ulva cpDNAs. Transfer RNA (tRNA) genes were searched for by reconstructing their cloverleaf structures using the tRNA scan-SE 1.21 software with default parameters2 (Schattner et al., 2005). Introns were determined and annotated by aligning the homologous host genes from the 23 Ulva chloroplast genomes (Table 1). Intron insertion-sites were identified based on the alignments of nucleotide (nt) sequences for the homologous genes with the counterparts in the chloroplast genome of U. compressa (MW353781) (Uco3) as reference. Intron name was defined as host gene plus insertion site. The class and core structure of all these introns were determined using the software RNAweasel (Lang et al., 2007) and Mfold (Zuker, 2003). The core domains of intron-encoded proteins (IEPs or intronic orfs) were determined by significant Pfam-A matches (Punta et al., 2012). Thus far, a total of 20 chloroplast genome sequences of Ulva species have been deposited in GenBank (Table 1), but some annotations in these data are incomplete or incorrect, which affects the accuracy of our comparative analysis. To solve this problem, we re-annotated all of the deposited Ulva chloroplast genomes in GenBank with the same method.
Phylogenetic Analysis of Intron-Encoded Proteins (IEPs) in cpDNAs and mtDNAs
So far, in Ulva organellar genomes (cpDNAs and mtDNAs), the LAGLIDADG homing endonuclease (LHE) as an intron-encoded protein (IEP) was detected in five types of introns including group IB (complete), group IA3, group I (derived, B2), group ID, and group II (LHE). To understand relationships of LHEs embedded in these five types of introns from Ulva cpDNAs and mtDNAs, phylogenetic analysis was performed based on the amino acid (aa) sequences of LAGLIDADG endonuclease domains. The LAGLIDADG endonuclease domains were searched from the aa sequences of the LHE proteins from these above five types of introns. The aa sequences of 139 LAGLIDADG endonuclease domain regions (64 in cpDNAs and 75 in mtDNAs) were subjected to concatenated alignments using ClustalX 1.83 with the default settings (Thompson et al., 1997). In most group IIA and IIB introns of Ulva chloroplast and mitochondrial genomes, the IEP was one reverse transcriptase/maturase (RTM). The conserved reverse transcriptase (RT) domains with relatively complete structure were searched from the aa sequences of these RTMs to analyze their relationships (Liu F. et al., 2020). The aa sequences of 67 RT domains (40 in cpDNAs and 27 in mtDNAs) from group IIA and IIB introns were subjected to concatenated alignments using ClustalX 1.83 with the default settings. Maximum Likelihood (ML) phylogenetic trees were constructed for LHE and RT datasets based on the Jones et al. (1992) w/freq. model with 1000 bootstrap replicates using the software MEGA 7.0 (Kumar et al., 2016). Initial tree(s) for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using a JTT model. There were a total of 338 and 309 positions in the final datasets of LHEs and RTs, respectively.
Comparative Genomic and Phylogenomic Analyses
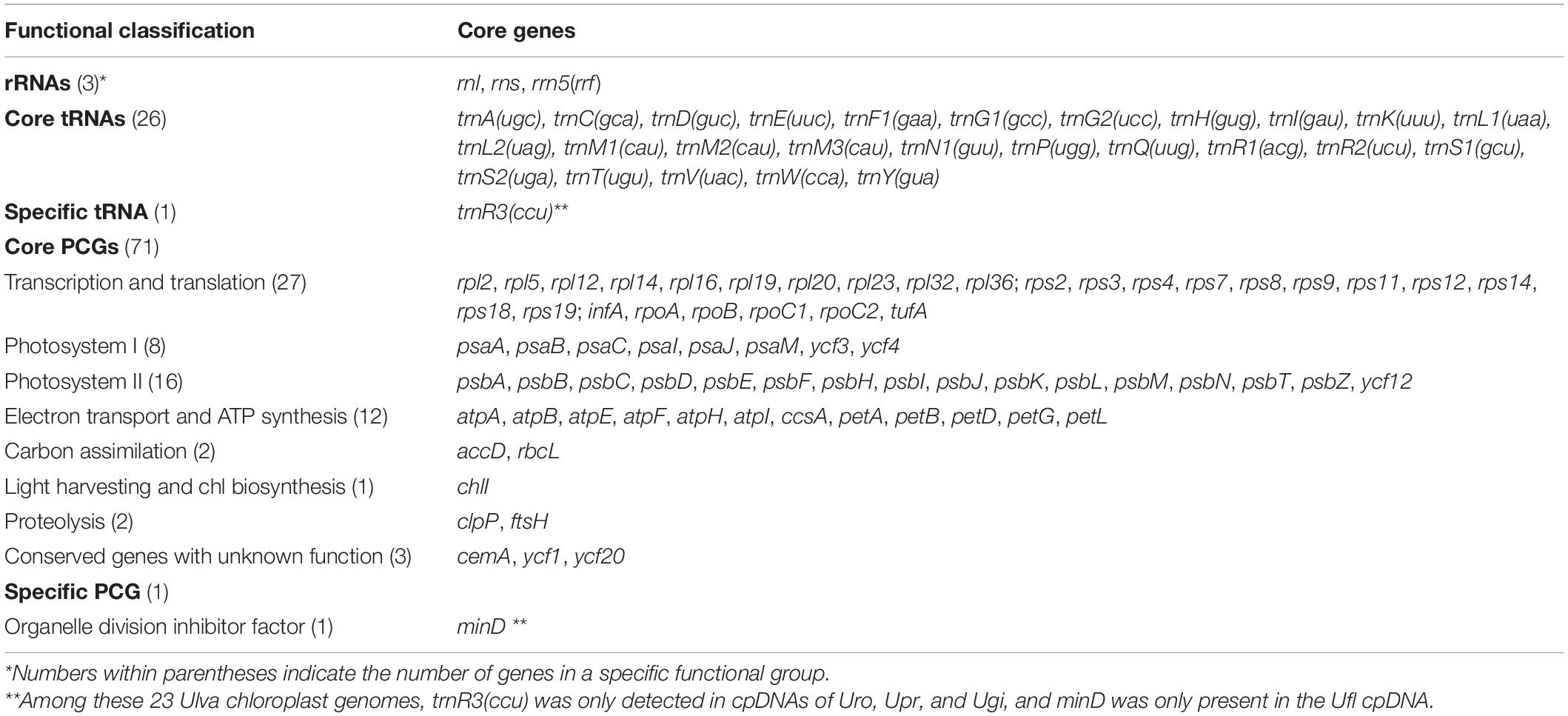
Base composition of the 23 Ulva cpDNAs was determined by the MEGA 7.0 software (Kumar et al., 2016). A total of 100 common genes including 71 PCGs, three rRNA genes, and 26 tRNA genes were shared among the 23 Ulva chloroplast genomes. Differences and identity percentages of the nucleotide (nt) sequences of these genes were evaluated using the BioEdit v7.1.9 software (Hall, 1999). The nt sequences of the 100 common genes and the aa sequences of the 71 PCGs were subjected to concatenated alignments using ClustalX 1.83 with the default settings, respectively (Thompson et al., 1997). For the nt dataset of the 100 conserved genes, the evolutionary history was inferred by using the Maximum Likelihood (ML) method based on the Tamura-Nei model (Tamura and Nei, 1993). Initial tree(s) for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach. For the aa dataset of the 71 PCGs, the evolutionary history was inferred by using the Maximum Likelihood method based on the Jones et al. w/freq. model (Jones et al., 1992). Initial tree(s) for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using a JTT model. All positions containing gaps and missing data were eliminated. There were a total of 66,558 and 19,985 positions in the final nt and aa datasets, respectively. Phylogenomic analysis was conducted with 1,000 bootstrap replicates using MEGA 7.0 (Kumar et al., 2016).
Results and Discussion
Features and Architecture of U. compressa Chloroplast Genomes
The three newly sequenced chloroplast genomes of U. compressa are 114,291 bp in Uco1, 91,189 bp in Uco2 and 96,824 bp in Uco3, respectively (Table 1), which are in the range of the known Ulva cpDNA size (86.7-119.9 kb) (Melton et al., 2015; Cai et al., 2017; Wang et al., 2017; Suzuki et al., 2018; Jiang et al., 2019; Fort et al., 2020). The variation in genome size of U. compressa cpDNAs at intraspecific level was mainly caused by differences in gain or loss of group I/II introns, integration of foreign DNA fragments, and content of non-coding intergenic spacer regions (Figure 1), which was similar to that observed in Ulva mitochondrial genomes (Liu F. et al., 2020). The chloroplast genomes of U. compressa tend to have high G + C content, ranging from 25.80% in Uco6 to 26.25% in Uco5, which is similar to that in Ulva rotundata (Uro) (26.12%), but richer than that in other Ulva species (23.91-25.73%) (Table 1). Only one overlapping region was detected in these seven U. compressa chloroplast genomes, which was the 17-bp psbD-psbC overlapping region (GTGGAAACGCTCTTTAA). This overlapping region was highly conserved in length and sequence in Ulvophyceae, even in Chlorophyta. Most of PCGs had a methionine (ATG) start codon in all sequenced Ulva cpDNAs, while psbC and infA started with GTG and TTG, respectively. Similar to that in most other Ulva speices, the rps19 was started with ATG in U. compressa, while it initiated with GTG in U. linza (Uli) and U. prolifera (Upr) (LP clade) (Liu et al., 2013).
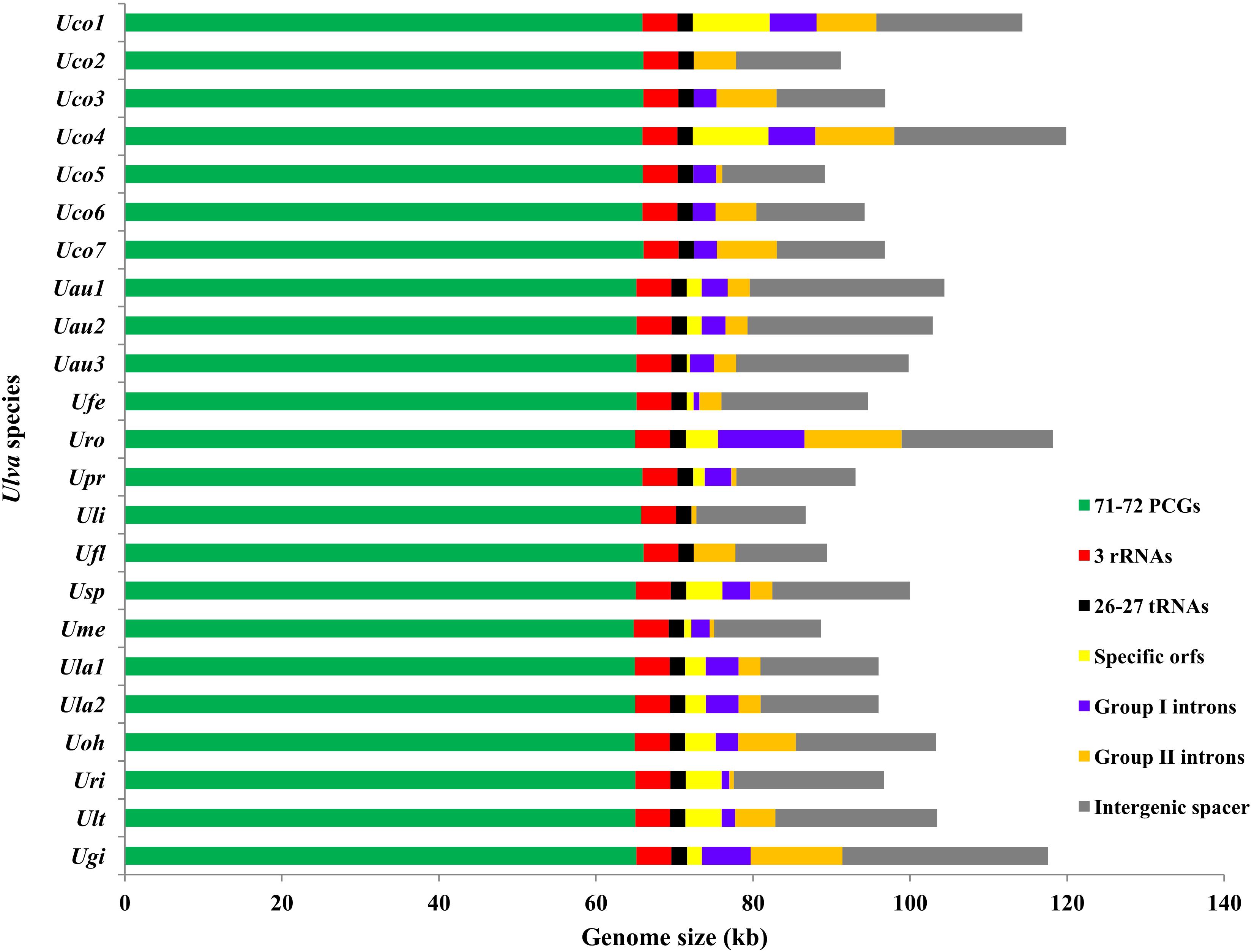
Figure 1. Contributions of conserved 71-72 PCGs, 3 rRNA genes (rRNAs), 26-27 tRNA genes (tRNAs), genome-specific free-standing orfs, group I introns, group II introns, and non-coding intergenic spacer regions in the 23 Ulva chloroplast genomes. The PCG proportion in Uco5 was adjusted due to the faulty assembly of some PCGs (rpoA, rpoB, rpoC1, and rpoC2). Horizontal bars represented the size of the Ulva cpDNAs.
Like other Ulva chloroplast genomes, the U. compressa cpDNAs do not have the quadripartite architecture and belong to IR-lacking chloroplast genomes (Figure 2). Chloroplast genomes usually harbor two identical copies of an inverted repeat (IR) region which carry the chloroplast rRNA operons. The IR region could pair for flip-flop recombination (Cattolico et al., 2008), and undergo expansion or contraction in different lineages of eukaryotic algae and land plants (e.g., Ruck et al., 2014; Liu et al., 2017a). The variability in IR size was often caused by the excision of sequences from the IR termini or by the integration of sequences in the adjacent small and large single-copy regions (Turmel and Lemieux, 2018). So far, the IR-containing ulvophycean chloroplast genomes have been detected in five orders including Ulvales (Pseudoneochloris marina), Ulotrichales (Pseudendoclonium akinetum, Trichosarcina mucosa, Hazenia capsulata, and Capsosiphon fulvescens), Oltmannsiellopsidales (Oltmannsiellopsis viridis and Dangemannia microcystis), Ignatiales (Pseudocharacium americanum and Ignatius tetrasporus), and Trentepohliales (Trentepohlia odorata), while IR-lacking chloroplast genomes have been found in three orders including Ulvales (Ulva spp.), Ulotrichales (Gloeotilopsis spp.), and Bryopsidales (all species), indicating that the IR has been lost multiple times during the evolution of ulvophycean green algae (Turmel et al., 2017).
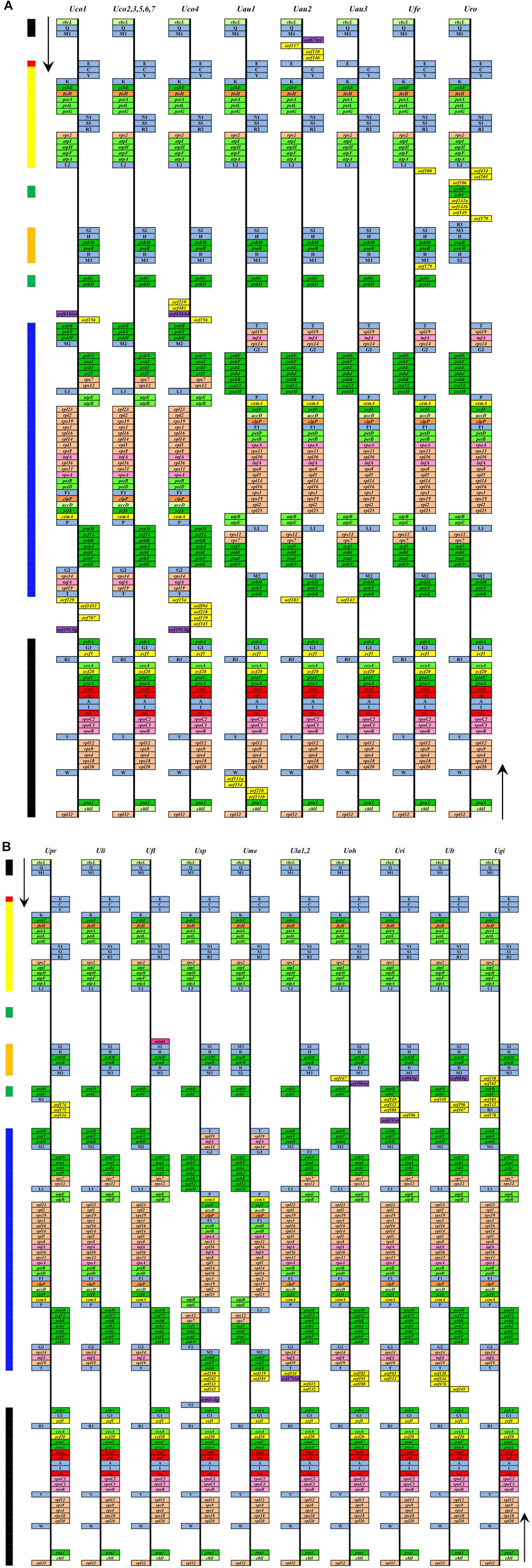
Figure 2. Comparison of genome organization and gene order of the Ulva chloroplast genomes in Ulva lineage II (A) and lineage I (B). Thick lines with different colors on the left represented different gene blocks. The arrows indicated the direction of gene transcription. Different gene types were shown as filled boxes in different colors.
Variation of Gene Repertoires in Ulva Chloroplast Genomes
The gene repertoires (including orfs) of U. compressa displayed marked variation at the intraspecific level, ranging from 102 genes in Uco2 to 120 genes in Uco4 (Figure 2). The variations of gene content among the Ulva chloroplast genomes were mainly caused by the different acquisitions of exogenous DNA fragments which usually carried specific free-standing orfs (over 100 codons), and the different gain or loss of group I/II introns which usually harbored intronic orfs, as was the same as that observed in the Ulva mitogenomes (Liu et al., 2017b; Liu F. et al., 2020).
Comparative analysis indicated that the sequenced 23 Ulva chloroplast genomes shared the same set of 100 conserved genes including 71 PCGs, three rRNA genes (rnl, rns, and rrn5), and 26 tRNA genes (Table 2). The organelle division inhibitor factor gene, minD, was lost in most Ulva chloroplast genomes, while it was situated between trnL2 and trnS2 only in the Ulva flexuosa (Ufl) cpDNA (Figure 2). This gene can be detected in chloroplast genomes of Ulvales (U. flexousa and P. marina), Ulotrichales, and Oltmannsiellopsidales (Pombert et al., 2005, 2006; Turmel et al., 2017; Kim et al., 2019), while it was lost in cpDNAs of Ulvales (most of Ulva spp.), Bryopsidales, Ignatiales, and Trentepohliales (Melton et al., 2015; Cremen et al., 2019; Zhu et al., 2019), indicating several independent losses in the different ulvophycean lineages. One specific tRNA gene, trnR3(ccu), which was located in the downstream neighborhood of psbC, was present only in U. rotundata (Uro), U. prolifera (Upr), and U. gigantea (Ugi). Considering that this gene is homologous in these three species, it is likely that this gene has been lost several times in the Ulva lineage. Two previously annotated tRNA genes, trnN2(auu) between psbB and psbA in Ulva sp. (Usp), and trnF2(aaa) between psbN and trnM2 in U. lactuca (KT882614) (Ula1) and Usp (Melton et al., 2015; Melton and Lopez-Bautista, 2017), did not display a standard cloverleaf structure, thus these two putative tRNA genes were not included here.
Genome-specific free-standing orfs were selectively distributed in some specific intergenic regions (e.g., trnM1-trnE, trnL2-psbD, psbC-trnM3, trnM3-psbD, psbB-psbD, psbC-psbB, trnT-psbA, psbA- psbB, and trnW-psaJ), in most of which genome rearrangement and recombination occurred more common among Ulva cpDNAs (Figure 2). Among these specific orfs, seven orfs displayed sequence similarity to the tyrosine-type recombinase/integrase (tri) of putative bacterial origin, belonging to DNA breaking-rejoining enzyme superfamily. Although these recombinases in Ulva cpDNAs showed great change in size from 67 to 619 aa, they shared the same fold in their C-terminal catalytic domain containing conserved active site residues. Five orfs ranging in size from 88 to 601 aa showed high similarity to the NAD-dependent DNA ligase (lig) of bacterial origin. The other specific orfs had little sequence similarity to any protein-coding genes in the GenBank database.
Some specific free-standing orfs are not shared among different individuals within the species (Figure 2). The block of tri-orf156 was present in Uco1 and Uco4, but not in Uco2, Uco3, and Uco5-7. The block of orf131a-orf154-orf210-orf131b was present only in U. australis (MN853875) (Uau1), while orf146-orf110-orf117-tri was present only in U. australis (LC507117) (Uau2). This shows that these genome-specific sequence regions were incorporated by recent horizontal gene transfer, and most importantly, invasion and integration of exogenous DNA fragments occurred frequently at intraspecific level of Ulva species. It has been known that chloroplast genomes can integrate foreign DNA sequences from diverse sources including mitochondrial sequences, bacteria, virus and jumping DNA/RNA-protein complex (Lang and Nedelcu, 2012; Straub et al., 2013; Turmel and Lemieux, 2018). In the cpDNAs of two siphonous green algae (Bryopsis plumosa and Tydemania expeditiones), the presence of bacterial genes with mobile functions (transposases, integrases, and phage/plasmid DNA primases) and bacterial DNA methyltransferases suggested that these genes had been acquired from bacteria through horizontal gene transfer (Leliaert and Lopez-Bautista, 2015). Short mtDNA fragments were present in two distinct regions of the Gloeotilopsis sarcinoidea cpDNA, providing the first evidence for intracellular inter-organelle gene migration in green algae (Turmel et al., 2016).
Diversity of Ulva Chloroplast Introns
The Ulva compressa chloroplast genomes harbored variable group I and II intron content from three in Uco2 to 11 in Uco4, occupying 5.92% of the cpDNA in Uco2 to 13.36% in Uco4. The variation in intron content was one of the most important factors responsible for genome size variation at intraspecific and interspecific levels in Ulva cpDNAs (Figure 1). Comparative analysis showed that a total of 26 chloroplast intron insertion-sites were detected in 14 host genes (atpA, atpB, atpI, infA, psaA, psaB, psbA, psbB, psbC, psbD, petB, petD, rnl, and rns) among the 23 Ulva cpDNAs (Table 3). With the exception of intron infA-62, almost all chloroplast introns displayed the idiosyncratic scattered distribution in the Ulva lineage, reflecting their frequent jumping and efficient dispersal as self-splicing and mobile genetic elements (Bonen and Vogel, 2001; Szitenberg et al., 2010). DNA sequences of both group I and II introns from the same insertion site were homologous among Ulva cpDNAs, as was the same as that observed in Ulva mtDNAs.
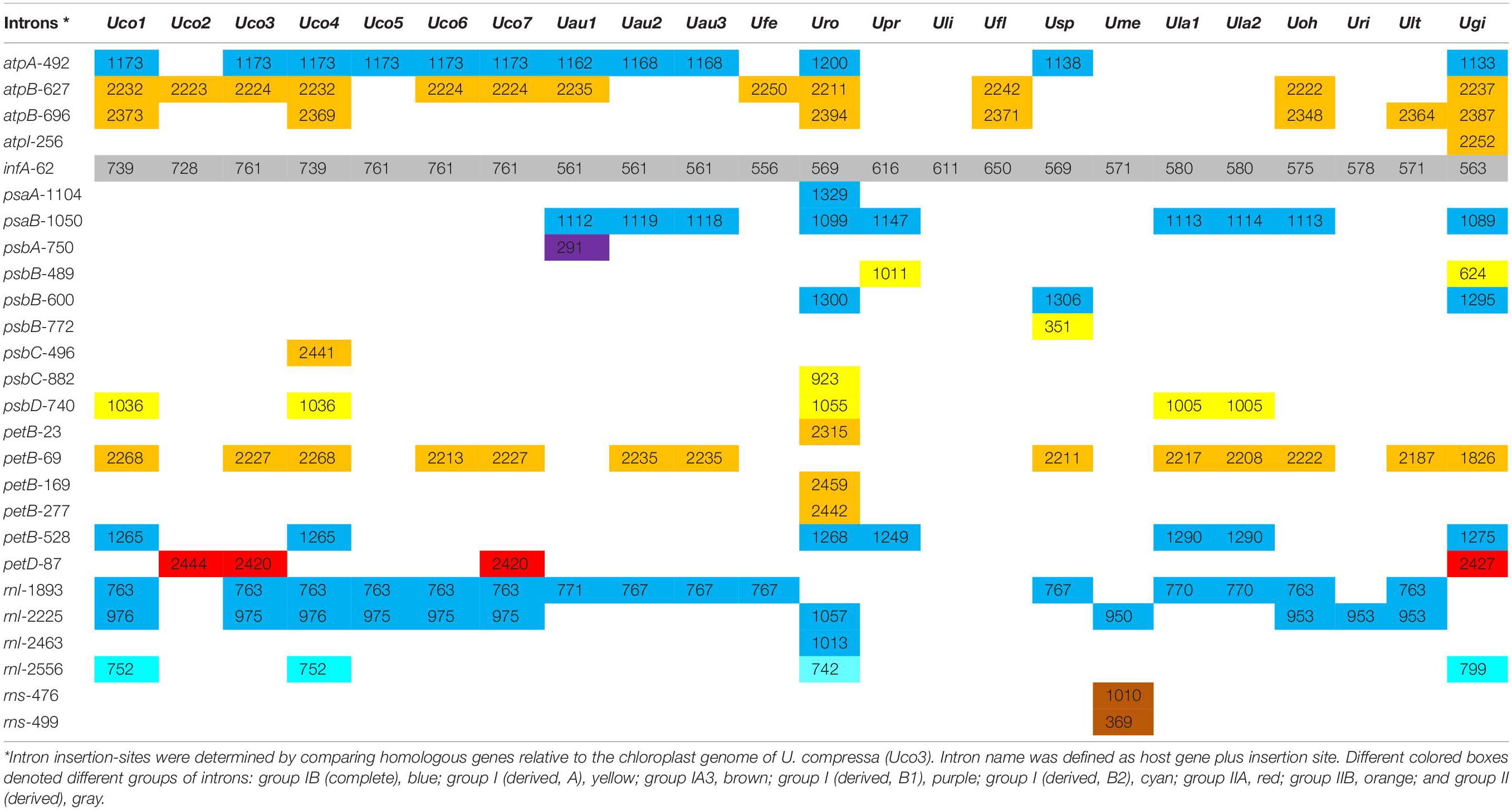
Table 3. Comparison of insertion site, size and group of introns in the 23 Ulva chloroplast genomes.
A total of 75 group I introns were observed among these 23 Ulva chloroplast genomes, and belonged to five types including group IB (complete), group I (derived, A), group IA3, group I (derived, B1), and group I (derived, B2) (Table 3). Among these Ulva cpDNAs, group IB (complete) introns were the most common, appearing at eight insertion sites. Approximately 90.7% of group I introns contained the intron-encoded proteins (IEPs) (over 100 codons) which could be classified into two families of endonucleases based on the presence of conserved amino acid motifs including LAGLIDADG and GIY-YIG (Haugen et al., 2005; Stoddard, 2011). Three degenerate introns including intron psbA-750, psbB-772 and rns-499 completely lost their IEPs. Most group I introns (85.3%) in Ulva cpDNAs contained a LAGLIDADG homing endonucleases (LHEs) with one or two LAGLIDADG motifs. The LHEs with only one motif, e.g., LHEs embedded in intron rnl-1893, work as homodimers that were limited to recognition of palindromic and near-palindromic target sites. The LHEs that possess two motifs in a single protein chain, e.g., LHEs in intron atpA-492 and psaB-1050, act as monomers that could target completely asymmetric invasion sites. Additionally, intron psbB-489, psbC-882 and psbD-740, which belonged to group I (derived, A), harbored a GIY-YIG homing endonuclease (GHE) (Table 3). Members of GHE family, which were usually found in bacteriophage (Sharma et al., 1992; VanRoey et al., 2002), displayed multidomain structures and recognized long non-palindromic sequences with significantly reduced fidelity (Landthaler et al., 2006). Differences in the homing endonuclease sequences indicated independent origins of LHE- and GHE-containing introns. Previously, we found that one group ID intron (intron cox1-709) was present in the clade of U. prolifera-U. linza-U. flexuosa-Ulva sp. (Upr-Uli-Ufl-Usp) in Ulva mtDNAs (Liu F. et al., 2020), however, no group ID intron was detected in the 23 Ulva cpDNAs.
Three types of group II introns were found among the 23 Ulva cpDNAs, including group IIA, group IIB, and group II (derived) (Table 3). The Ulva cpDNAs contained more group IIB introns detected at eight insertion sites than group IIA introns at one insertion site. Previously, group II introns which carried a putative LAGLIDADG homing endonuclease (LHE) were present in Ulva mtDNAs, while none was found in Ulva cpDNAs. Considering the intron petD-87 was only found in Uco2, Uco3, Uco7 and Ugi, it is likely that this intron was obtained by recent horizontal transfer. The IEP in group IIA and IIB introns was one multifunctional protein with reverse transcriptase/maturase (RTM), and DNA endonuclease (ENase) activities in addition to their ribozyme component, which catalyzes splicing (Dai et al., 2003; Lambowitz and Zimmerly, 2011). Reverse transcriptase (RT) plays a key role in the mobility of organellar group II introns, and maturase aid in intron splicing (Seetharaman et al., 2006; Novikova and Belfort, 2017).
The infA gene harbored one group II (derived) intron (intron infA-62) in all of the sequenced Ulva cpDNAs (Table 3), which has not been detected and annotated before. The intron infA-62 in U. compressa ranged in size from 728 bp in Uco2 to 761 bp in Uco3, and Uco5-7, which were larger than that in other Ulva species (556-650 bp) (Table 4). This intron was degenerate in all these Ulva cpDNAs and its intronic orf was completely lost, whereas the sequences of splicing sites and domain V were highly conserved. However, this intron was absent in the closest neighbor of Ulva species, Pseudoneochloris marina (Ulvaceae), and the other chlorophycean taxa, indicating that it should be the result of an independent invasion event occurred in a common ancestor of Ulva species by horizontal transfer.
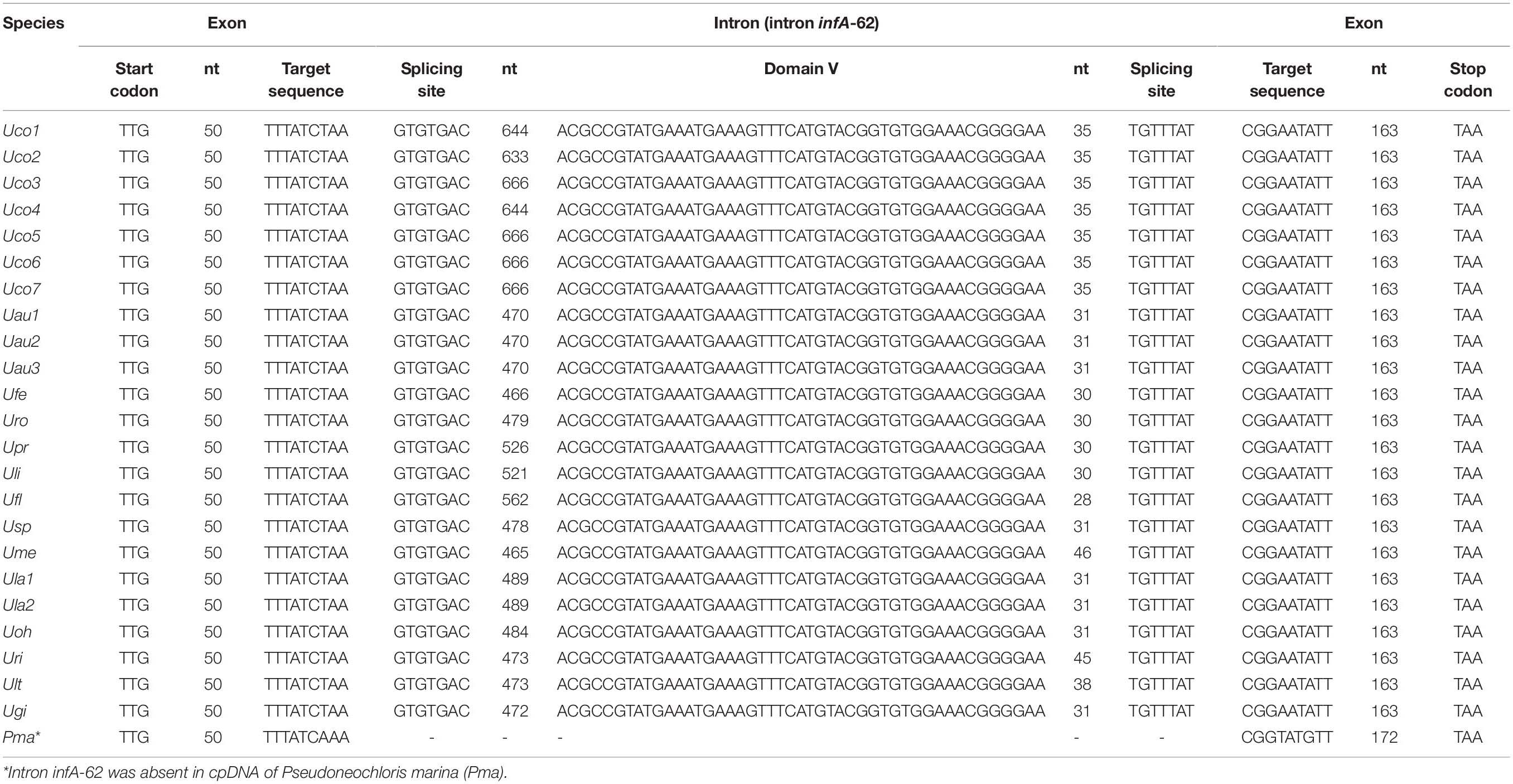
Table 4. One group II (derived) intron was identified in the infA genes of all sequenced Ulva chloroplast genomes.
Phylogenetic Analysis of Chloroplast and Mitochondrial IEPs (LHEs and RTMs)
The LHEs were embedded in three types of group I introns among Ulva cpDNAs, including group IB (complete) (at eight insertion sites), group IA3 (at one), and group I (derived, B2) (at one) (Table 3), whereas among Ulva mtDNAs, they were embedded in three types of introns including group IB (complete) (at eight), group ID (at one), and group II (LHE) (at five) (Liu F. et al., 2020). To further understand relationships of these intronic LHEs from chloroplast and mitochondrial genomes in Ulva species, a ML tree was constructed based on the aa sequences of the LAGLIDADG endonuclease domain regions from all LHEs in Ulva cpDNAs and mtDNAs. Most obviously, the LHEs from cognate introns clustered together first (Figure 3 and Supplementary Figure 1), indicating that they originated from their common ancestor. Second, some LHEs in chloroplast group IB (complete) introns (intron atpA-492 and petB-528) show high homology to that in mitochondial introns (intron atp1-540), indicating that these group IB (complete) introns should be of the same origin, and migrate or expand in intracellular inter-organelle genomes. Some group II (LHE) introns (e.g., intron rnl-2698 and rns-420) have been observed to propagate and expand within Ulva mitochondrial genomes (e.g., U. flexuosa and U. linza). Finally, the mitochondrial LHEs in group IB (complete) introns have close relationships with that in group II (LHE) introns, e.g., intron cox1-734 vs rnl-2698/rns-420, and intron cox1-281 vs rnl-2080/rns-670 (Figure 3), although these two types of related introns show great difference in ribozyme components.
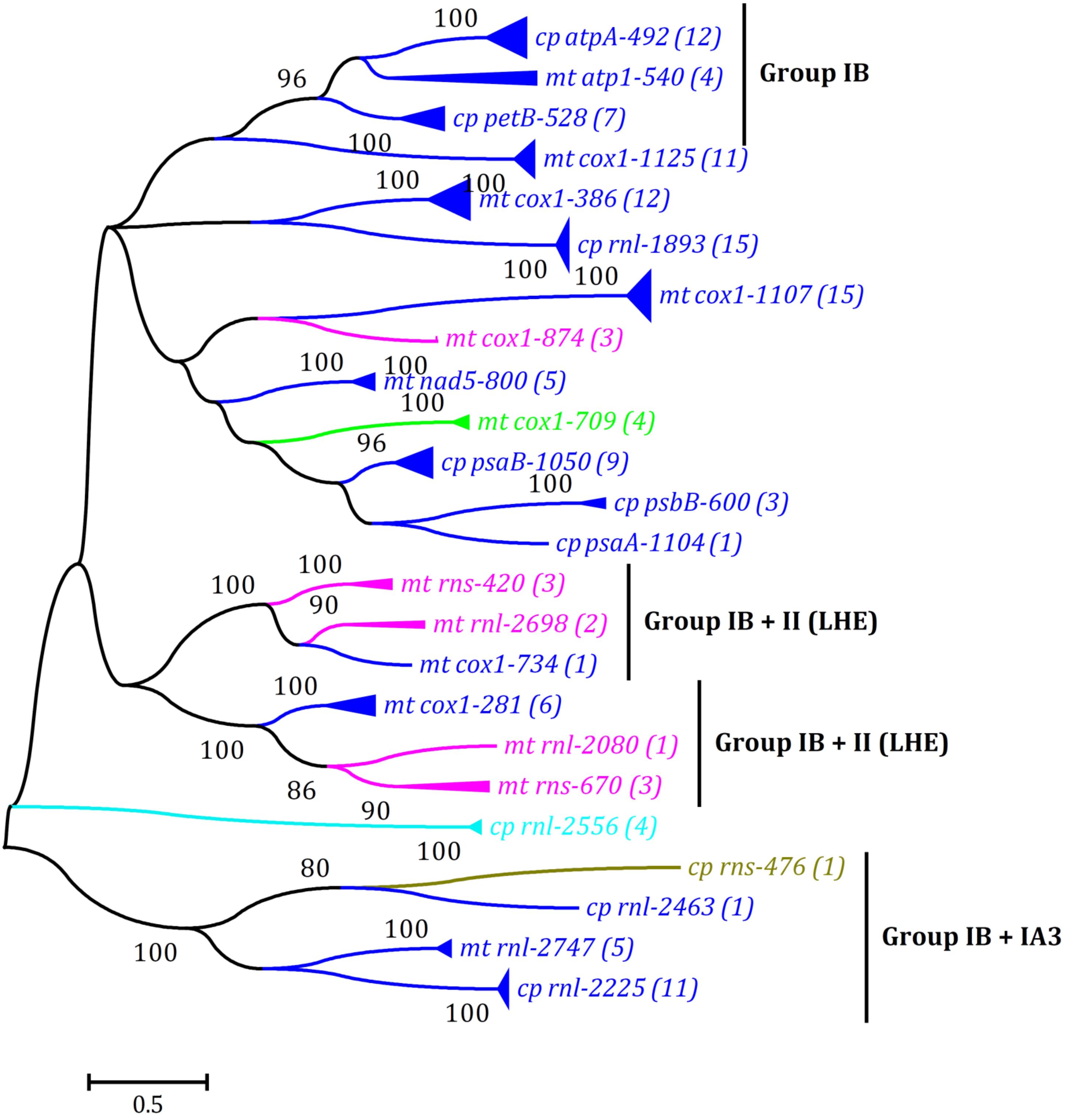
Figure 3. Phylogenetic tree based on Maximum Likelihood (ML) analysis of amino acid (aa) sequences of 139 LAGLIDADG endonuclease domain regions (64 in cpDNAs and 75 in mtDNAs) from five types of introns including group IB (complete), group IA3, group I (derived, B2), group ID, and group II (LHE). Different colored lines denoted LAGLIDADG endonuclease domains from different groups of introns: group IB (complete), blue; group IA3, brown; group I (derived, B2), cyan; group ID, green; and group II (LHE), pink. The ML analysis was conducted with 1,000 bootstrap replicates using MEGA 7.0. The bootstrap support values greater than 80% were displayed at branches. Branch lengths were proportional to the amount of sequence change, which were indicated by the scale bar below the trees. The numbers in parentheses represented the number of LHE proteins from cognate introns. The complete phylogenetic tree was shown in Supplementary Figure 1.
The ML phylogenetic tree was built based on the aa sequences of RT domains from all RTMs in group IIA and IIB introns of Ulva cpDNAs and mtDNAs. Similar to the LHE tree, the RTs from cognate introns were grouped together first, revealing their close relationships. The distinctness of the two RT lineages (group IIA and IIB) was well supported by high bootstrap values (100%) (Figure 4 and Supplementary Figure 2). The chloroplast and mitochondrial RTMs coevolved with their associated intron RNA structure (ribozyme components) in the genus Ulva. In some Ulva chloroplast genomes, the RTs in different group IIB introns are of the same origin, e.g., those in intron atpB-627 and petB-69 of Uco1, Uco3, Uco4, Uco6, Uco7, Uoh and Ugi, and those in intron petB-169 and petB-277 of Uro (Figure 4), indicating that these introns are likely to have experienced proliferation and expansion within these Ulva chloroplast genomes.
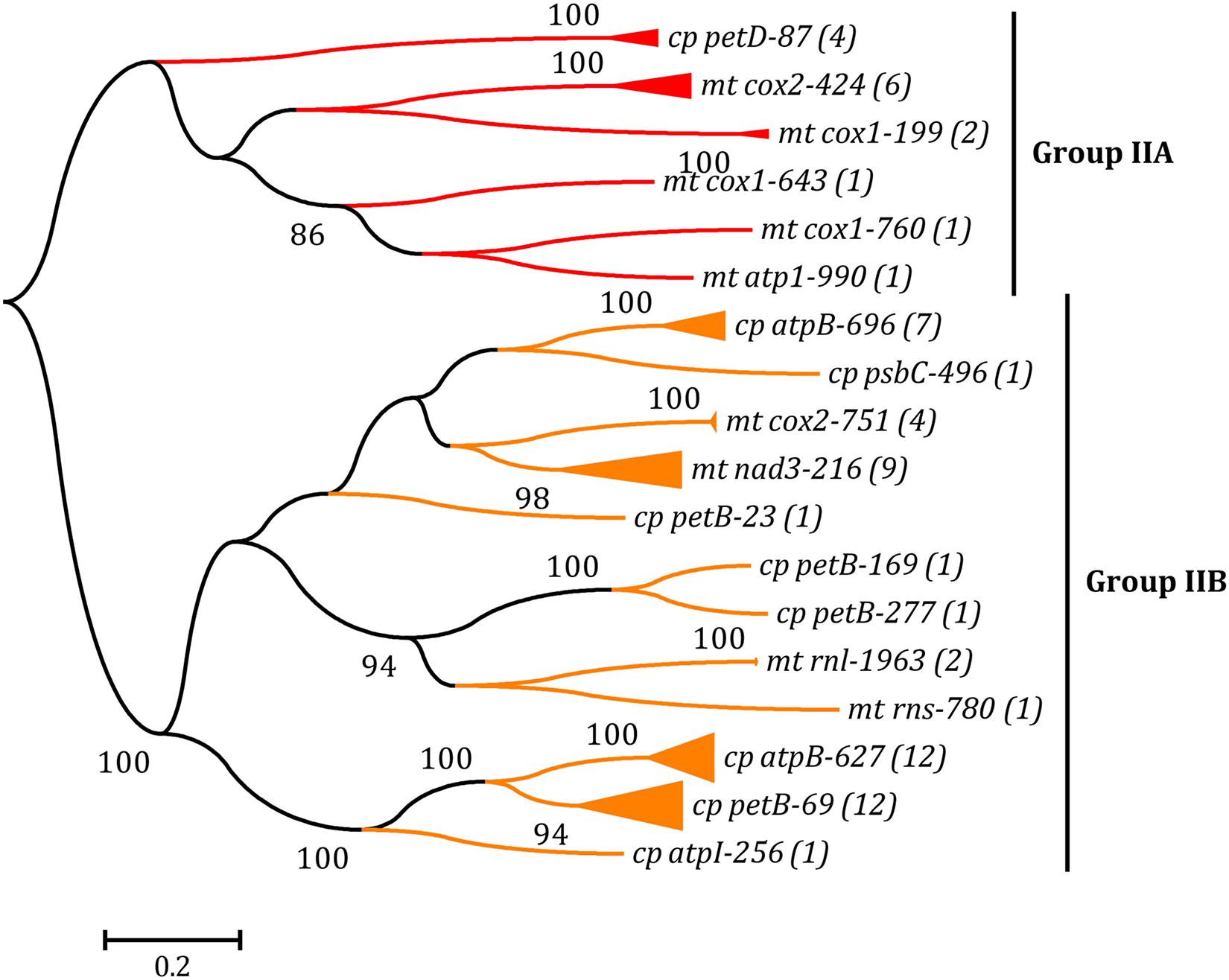
Figure 4. Phylogenetic tree based on Maximum Likelihood (ML) analysis of amino acid (aa) sequences of 67 reverse transcriptase (RT) domains (40 in cpDNAs and 27 in mtDNAs) from group IIA and IIB introns. The ML analysis was conducted with 1,000 bootstrap replicates using MEGA 7.0. The bootstrap support values greater than 80% were displayed at branches. Branch lengths were proportional to the amount of sequence change, which were indicated by the scale bar below the trees. The numbers in parentheses represented the number of RTM proteins from cognate introns. The complete phylogenetic tree was shown in Supplementary Figure 2.
Integrated Analysis of Rearrangement Events and Phylogenomic Relationships
To understand the evolution of chloroplast genome architecture in the genus Ulva, integrated analysis was performed by combining gene orders of these 23 Ulva cpDNAs with phylogenomic relationships of these Ulva species. The Ulva chloroplast genomes displayed intrageneric variability in genome structure (Figure 2), although their core gene repertoires were highly conserved (Table 2). The seven U. compressa cpDNAs displayed the same genome organization, representing a novel gene order which was different from that of other Ulva cpDNAs due to multiple rearrangement events (Figure 2). To examine the phylogenomic relationships among these Ulva species, the Maximum Likelihood (ML) trees were constructed on the bases of the nt dataset of 100 common genes and the aa dataset of 71 common PCGs, respectively. The topology of the two ML trees were very similar, and both trees showed that the 23 Ulva cpDNAs from 14 species were clustered into two large clades with high support values (100% bootstrap), representing two Ulva lineages, I and II (Figure 5), as was similar to our previous finding based on the nt dataset of 61 mitochondrial common genes (Liu F. et al., 2020). Based on a comparative analysis of the aligned nt sequences of 100 common chloroplast genes including 71 PCGs, 3 rRNAs, and 26 tRNAs, the sequence identity values at intraspecific level in U. compressa ranged from 98.6% (different in 965 bp) to 99.9% (different in 51 bp) (Table 5). The maximum values of intraspecific divergences in U. compressa even exceeded that of some interspecific divergences, e.g., U. prolifera (Upr) vs U. linza (Uli), and U. rigida (Uri) vs U. laetevirens (Ult). It is a remarkable fact that inter-species hybridization between Upr and Uli (Hiraoka et al., 2011, 2017) and between Uri and Ult (Fort et al., 2020) has been observed to some extent, indicating that they are genetically closely related species. These evidences challenge us to understand species concepts and species boundaries in the genus Ulva, and especially in the U. compressa clade.
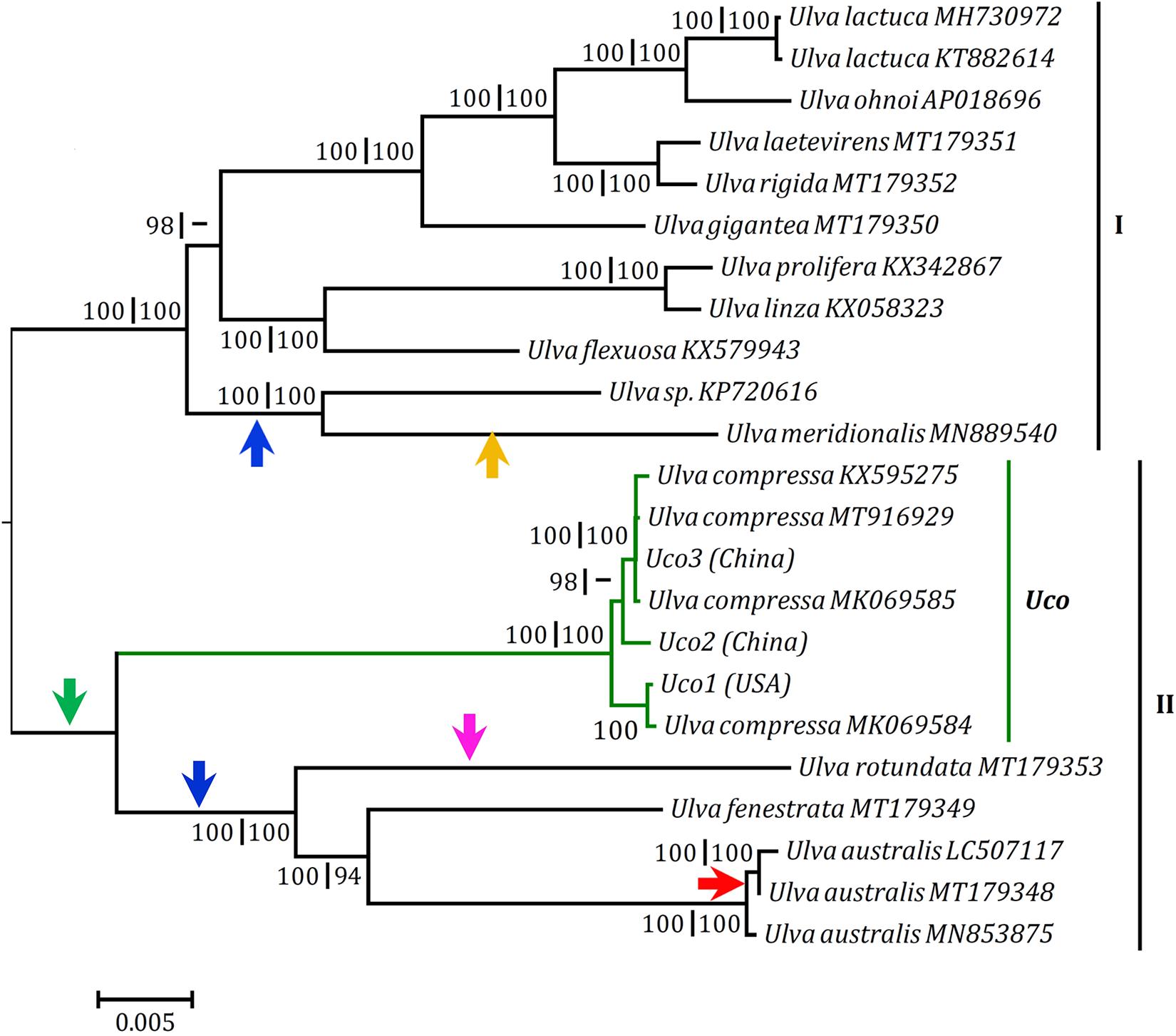
Figure 5. Phylogenomic trees based on Maximum Likelihood (ML) analysis of the nucleotide (nt) sequences of the 100 common genes and the amino acid (aa) sequences of the 71 common PCGs in the 23 Ulva chloroplast genomes. The ML analysis was conducted with 1,000 bootstrap replicates using MEGA 7.0. The bootstrap support values greater than 80% were displayed at branches. Branch lengths were proportional to the amount of sequence change, which were indicated by the scale bar below the trees. Arrows with different colors represented reversal events of different gene blocks: two genes (psbD-psbC), green; 45 genes from psbB to trnT(ugu), blue; nine genes (psbD-psbC-trnR3-trnM3-trnD-psaB-psbM-trnH-trnS2), pink; six genes (trnM3-trnD-psaB-psbM-trnH-trnS2), orange; and trnE(uuc), red.
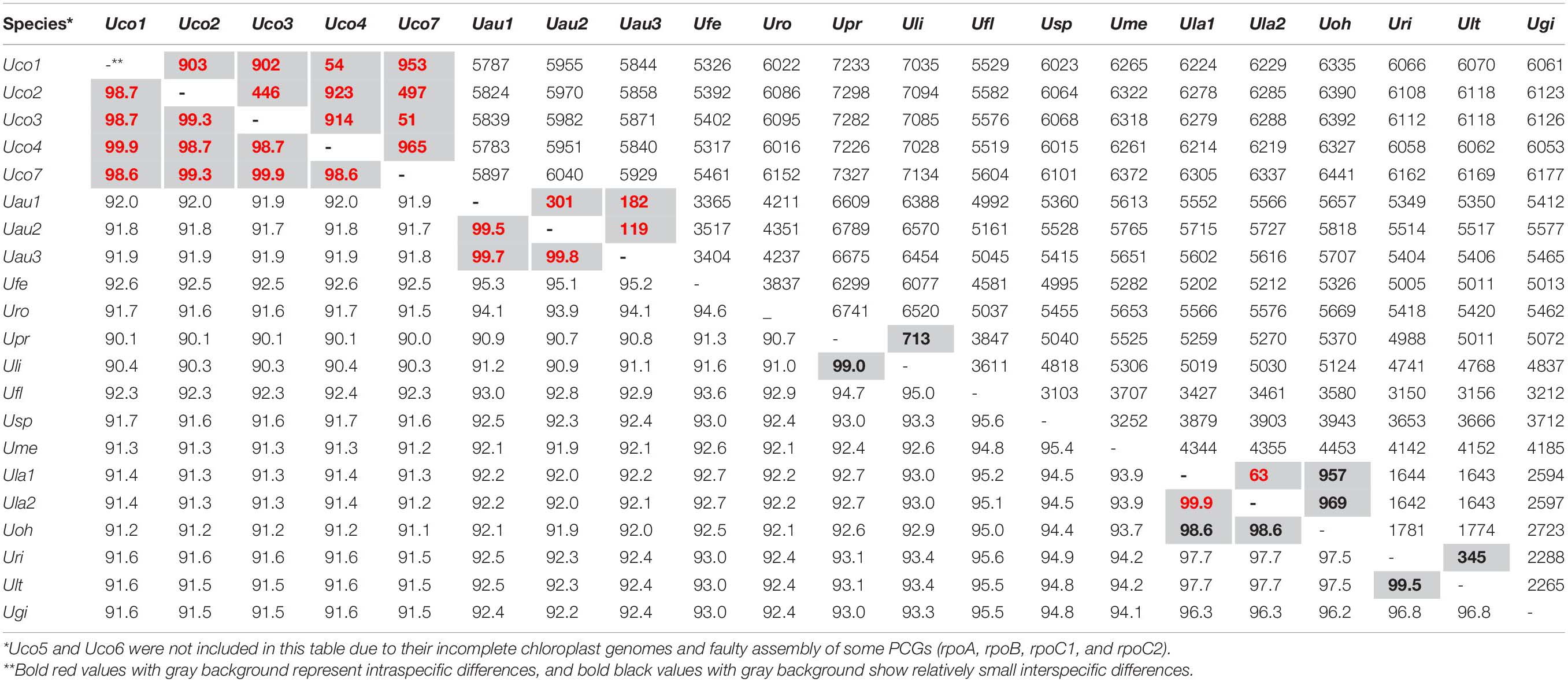
Table 5. Differences (bp, upper-right) and identity values (%, bottom-left) of the aligned nucleotide (nt) sequences of 100 common genes (71 PCGs, 3 rRNAs, and 26 tRNAs) from 21 complete chloroplast genomes of Ulva species.
A locally collinear block of two genes (psbD-psbC) has been observed to be inverted in cpDNAs of Ulva lineage II (Uco, Uau, Ufe, and Uro) (Figure 2). Considering the phylogenomic relationships of Ulva species, this rearrangement is likely to occur in the common ancestor of species in Ulva lineage II, after its divergence from lineage I (Figure 5). However, a collinear block of nine genes (psbD-psbC-trnR3-trnM3-trnD-psaB-psbM-trnH-trnS2) was reversed only in Uro, as probably occurred after the reversal of the psbD-psbC block in Ulva lineage II. A large gene block harboring 45 genes from psbB to trnT(ugu) (Melton et al., 2015; Melton and Lopez-Bautista, 2017) has been inverted in cpDNAs of the Ulva sp.-U. meridionalis (Usp-Ume) clade in Ulva lineage I, and cpDNAs of the U. australis-U. fenestrata-U. rotundata (Uau-Ufe-Uro) clade in lineage II, indicating that the rearrangement of this gene block should be two independent inversion events. Only in the Ume cpDNA is the reversal of a collinear block of six genes (trnM3-trnD-psaB-psbM-trnH-trnS2) observed, indicating that it should be a independent rearrangement event occurring after its divergence from Usp. A reversal event related to only a tRNA gene, trnE(uuc), happened in cpDNAs of Uau2 and Uau3, not in Uau1 and other Ulva cpDNAs, indicating that it was a recent rearrangement at intraspecific level. Compared with PCGs and/or rRNA genes, tRNA genes show more mobility in organellar genomes of eukaryotic algae (e.g., brown algae, diatoms, red algae) (e.g., Ruck et al., 2014; Liu et al., 2017a, 2019). Previously, intraspecific rearrangement was observed to occur in mitochondrial genomes of U. compressa. A collinear block of eight genes (rps11-rps19-rps4-rpl16-trnR-trnQ-trnE-trnS) with the size of 3,631 bp was inverted only in Uco1 mitogenome (Liu F. et al., 2020). Compared with the gene content and gene sequence, the structure of Ulva chloroplast genome is not conserved, but remarkably plastic, due to multiple rearrangement events. The observed multiple rearrangements at intraspecific and interspecific levels depict the structural diversity and evolutionary trend of chloroplast genomes in Ulva species.
Conclusion
This study on Ulva chloroplast genomes is the most extensive investigation of chlorophyte cpDNAs at the intragenus level. Our study illustrated the remarkable plasticity of Ulva chloroplast genomes among congeneric species, even within the species (e.g., U. compressa and U. australis). The variation in genome size at intraspecific and interspecific levels was mainly caused by differences in gain or loss of group I/II intron, integration of foreign DNA fragments, and content of non-coding intergenic spacer regions. The Ulva chloroplast genomes shared the same set of 100 conserved genes, however, the minD gene was detected only in Ulva flexuosa cpDNA. Five types of group I introns, most of which carry a LAGLIDADG or GIY-YIG homing endonuclease (LHE and GHE), and three of group II introns, usually encoding a reverse transcriptase/maturase (RTM), were detected at 26 insertion sites of 14 host genes in these Ulva chloroplast genomes, and many intron insertion-sites have been found for the first time in Chlorophyta. It is worth noting that one degenerate group II intron previously ignored has been observed in the infA genes of Ulva species, but not in the other chlorophycean taxa, indicating that it should be the result of an independent invasion event occurred in a common ancestor of Ulva species. The structure of Ulva chloroplast genomes is not conserved, but remarkably plastic, due to multiple rearrangement events. The present study provides important information to understand the evolution patterns of Ulva chloroplast genomes, and have important implications to understand molecular species concepts and species boundaries in the genus Ulva. Additional genomic data from other Ulva species are still needed to further decipher the mechanisms responsible for diversity and evolution of Ulva cpDNAs.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: MW548841, MW344287, and MW353781).
Author Contributions
FL designed the study, performed the analysis, and wrote the manuscript. Both authors performed the experiments have read and approved the final version of the manuscript.
Funding
This work was supported by the Science and Technology Basic Resources Investigation Program of China (No. 2018FY100200), the Key Research Program of Frontier Sciences, Chinese Academy of Sciences (No. QYZDB-SSW-DQC023), the Strategic Priority Research Program of Chinese Academy of Sciences (No. XDA23050302/XDA23050403), the Major Scientific and Technological Innovation Project of Shandong Province (No. 2019JZZY020706), the National Natural Science Foundation of China (No. 41876165), and the National Science Foundation (NSF) HBCU-UP (No. 1436759) for Postdoctoral Fellowship.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank Wei Luan, Yu Wang, Zhe Jin, Xingfeng Liu, Manman Liu, and Shitao Shi for their assistance in algal collection and data analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.668542/full#supplementary-material
Supplementary Figure 1 | Phylogenetic tree based on Maximum Likelihood (ML) analysis of amino acid (aa) sequences of 139 LAGLIDADG endonuclease domain regions (64 in cpDNAs and 75 in mtDNAs) from five types of introns including group IB (complete), group IA3, group I (derived, B2), group ID, and group II (LHE). Different colored lines denoted LAGLIDADG endonuclease domains from different groups of introns: group IB (complete), blue; group IA3, brown; group I (derived, B2), cyan; group ID, green; and group II (LHE), pink. The ML analysis was conducted with 1,000 bootstrap replicates using MEGA 7.0. The bootstrap support values greater than 80% were displayed at branches. Branch lengths were proportional to the amount of sequence change, which were indicated by the scale bar below the trees.
Supplementary Figure 2 | Phylogenetic tree based on Maximum Likelihood (ML) analysis of amino acid (aa) sequences of 67 reverse transcriptase (RT) domains (40 in cpDNAs and 27 in mtDNAs) from group IIA and IIB introns. The ML analysis was conducted with 1,000 bootstrap replicates using MEGA 7.0. The bootstrap support values greater than 80% were displayed at branches. Branch lengths were proportional to the amount of sequence change, which were indicated by the scale bar below the trees.
Footnotes
References
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Balar, N. B., and Mantri, V. A. (2020). Insights into life cycle patterns, spore formation, induction of reproduction, biochemical and molecular aspects of sporulation in green algal genus Ulva: implications for commercial cultivation. J. Appl. Phycol. 32, 473–484. doi: 10.1007/s10811-019-01959-7
Blomster, J., Bäck, S., Fewer, D. P., Kiirikki, M., Lehvo, A., Maggs, C. A., et al. (2002). Novel morphology in Enteromorpha (Ulvophyceae) forming green tides. Am. J. Bot. 89, 1756–1763. doi: 10.3732/ajb.89.11.1756
Blomster, J., Maggs, C. A., and Stanhope, M. J. (1998). Molecular and morphological analysis of Enteromorpha intestinalis and E. compressa (Chlorophyta) in the British Isles. J. Phycol. 34, 319–340. doi: 10.1046/j.1529-8817.1998.340319.x
Bonen, L., and Vogel, J. (2001). The ins and outs of group II introns. Trends Genet. 17, 322–331. doi: 10.1016/s0168-9525(01)02324-1
Cai, C., Wang, L., Zhou, L., He, P., and Jiao, B. (2017). Complete chloroplast genome of green tide algae Ulva flexuosa (Ulvophyceae, Chlorophyta) with comparative analysis. PLoS One 12:e0184196. doi: 10.1371/journal.pone.0184196
Cattolico, R. A., Jacobs, M. A., Zhou, Y., Chang, J., Duplessis, M., Lybrand, T., et al. (2008). Chloroplast genome sequencing analysis of Heterosigma akashiwo CCMP452 (West Atlantic) and NIES293 (West Pacific) strains. BMC Genomics 9:211. doi: 10.1186/1471-2164-9-211
Cocquyt, E., Verbruggen, H., Leliaert, F., and De Clerck, O. (2010). Evolution and cytological diversification of the green seaweeds (Ulvophyceae). Mol. Biol. Evol. 27, 2052–2061. doi: 10.1093/molbev/msq091
Cremen, M. C. M., Leliaert, F., Marcelino, V. R., and Verbruggen, H. (2018). Large diversity of non-standard genes and dynamic evolution of chloroplast genomes in siphonous green algae (Bryopsidales, Chlorophyta). Genome Biol. Evol. 10, 1048–1061. doi: 10.1093/gbe/evy063
Cremen, M. C. M., Leliaert, F., West, J., Lam, D. W., Shimada, S., Lopez-Bautista, J. M., et al. (2019). Reassessment of the classification of bryopsidales (chlorophyta) based on chloroplast phylogenomic analyses. Mol. Phylogenet. Evol. 130, 397–405. doi: 10.1016/j.ympev.2018.09.009
Dai, L., Toor, N., Olson, R., Keeping, A., and Zimmerly, S. (2003). Database for mobile group II introns. Nucleic Acids Res. 31, 424–426. doi: 10.1093/nar/gkg049
De Clerck, O., Kao, S. M., Bogaert, K. A., Blomme, J., Foflonker, F., Kwantes, M., et al. (2018). Insights into the Evolution of Multicellularity from the Sea Lettuce Genome. Curr. Biol. 28, 2921–2933.
Del Cortona, A., Leliaert, F., Bogaert, K. A., Turmel, M., Boedeker, C., Janouškovec, J., et al. (2017). The plastid genome in Cladophorales green algae is encoded by hairpin chromosomes. Curr. Biol. 27, 3771–3782. doi: 10.1016/j.cub.2017.11.004
Fort, A., McHale, M., Cascella, K., Potin, P., Usadel, B., Guiry, M. D., et al. (2020). Foliose Ulva species show considerable inter-specific genetic diversity, low intra-specific genetic variation, and the rare occurrence of inter-specific hybrids in the wild. J. Phycol. 57, 219–233. doi: 10.1111/jpy.13079
Fučíková, K., Leliaert, F., Cooper, E. D., Škaloud, P., D’Hondt, S., De Clerck, O., et al. (2014). New phylogenetic hypotheses for the core Chlorophyta based on chloroplast sequence data. Front. Ecol. Evol. 2:63. doi: 10.3389/fevo.2014.00063
Gao, G., Zhong, Z. H., Zhou, X. H., and Xu, J. T. (2016). Changes in morphological plasticity of Ulva prolifera under different environmental conditions: a laboratory experiment. Harmful Algae 59, 51–58. doi: 10.1016/j.hal.2016.09.004
Guiry, M. D., and Guiry, G. M. (2021). AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. Am. J. Plant Sci. 10:10.
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41, 95–98.
Haugen, P., Simon, D. M., and Bhattacharya, D. (2005). The natural history of group I introns. Trends Genet. 21, 111–119. doi: 10.1016/j.tig.2004.12.007
Hayden, H. S., and Waaland, J. R. (2002). Phylogenetic systematics of the Ulvaceae (Ulvales, Ulvophyceae) using chloroplast and nuclear DNA sequences. J. Phycol. 38, 1200–1212. doi: 10.1046/j.1529-8817.2002.01167.x
Hiraoka, M., Ichihara, K., Zhu, W., Ma, J., and Shimada, S. (2011). Culture and hybridization experiments on an Ulva clade including the Qingdao strain blooming in the Yellow Sea. PLoS One 6:e19371. doi: 10.1371/journal.pone.0019371
Hiraoka, M., Ichihara, K., Zhu, W., Shimada, S., Oka, N., Cui, J., et al. (2017). Examination of species delimitation of ambiguous DNA-based Ulva (Ulvophyceae, Chlorophyta) clades by culturing and hybridisation. Phycologia 56, 517–532. doi: 10.2216/16-109.1
Hughey, J. R., Maggs, C. A., Mineur, F., Jarvis, C., Miller, K. A., Shabaka, S. H., et al. (2019). Genetic analysis of the Linnaean Ulva lactuca (Ulvales, Chlorophyta) holotype and related type specimens reveals name misapplications, unexpected origins, and new synonymies. J. Phycol. 55, 503–508. doi: 10.1111/jpy.12860
Jiang, T., Gu, K., Wang, L., Liu, Q., Shi, J., Liu, M., et al. (2019). Complete chloroplast genome of Ulva prolifera, the dominant species of green macroalgal blooms in Yellow Sea, China. Mitochondrial. DNA Part B 4, 1930–1931. doi: 10.1080/23802359.2019.1610090
Jones, D. T., Taylor, W. R., and Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282. doi: 10.1093/bioinformatics/8.3.275
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kim, D., Lee, J. M., Choi, J. W., and Yang, J. H. (2019). Flip-flop organization in the chloroplast genome of Capsosiphon fulvescens (Ulvophyceae, Chlorophyta). J. Phycol. 55, 214–223. doi: 10.1111/jpy.12811
Kimura, M. (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/bf01731581
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lambowitz, A. M., and Zimmerly, S. (2011). Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb. Perspect. Biol. 3:a003616. doi: 10.1101/cshperspect.a003616
Landthaler, M., Shen, B. W., Stoddard, B. L., and Shub, D. A. (2006). I-BasI and I-HmuI: two phage intron-encoded endonucleases with homologous DNA recognition sequences but distinct DNA specificities. J. Mol. Biol. 358, 1137–1151. doi: 10.1016/j.jmb.2006.02.054
Lang, B. F., Laforest, M.-J., and Burger, G. (2007). Mitochondrial introns: a critical view. Trends Genet. 23, 119–125. doi: 10.1016/j.tig.2007.01.006
Lang, B. F., and Nedelcu, A. M. (2012). “Plastid genomes of algae,” in Advances in Photosynthesis and Respiration Including Bioenergy and Related Processes: Genomics of Chloroplasts and Mitochondria, eds R. Bock and V. Knoop (Dordrecht: Springer), 59–87. doi: 10.1007/978-94-007-2920-9_3
Leliaert, F., and Lopez-Bautista, J. M. (2015). The chloroplast genomes of Bryopsis plumosa and Tydemania expeditiones (Bryopsidales, Chlorophyta): compact genomes and genes of bacterial origin. BMC Genom. 16:204. doi: 10.1186/s12864-015-1418-3
Leliaert, F., Smith, D. R., Moreau, H., Herron, M. D., Verbruggen, H., Delwiche, C. F., et al. (2012). Phylogeny and molecular evolution of the green algae. Crit. Rev. Plant Sci. 31, 1–46.
Liu, F., Jin, Z., Wang, Y., Bi, Y., and Melton, J. T. (2017a). Plastid genome of Dictyopteris divaricata (Dictyotales, Phaeophyceae): understanding the evolution of plastid genomes in brown algae. Mar. Biotech. 19, 627–637. doi: 10.1007/s10126-017-9781-5
Liu, F., Melton, J. T., and Bi, Y. (2017b). Mitochondrial genomes of the green macroalga Ulva pertusa (Ulvophyceae, Chlorophyta): novel insights into the evolution of mitogenomes in the Ulvophyceae. J. Phycol. 53, 1010–1019. doi: 10.1111/jpy.12561
Liu, F., Melton, J. T., Lopez-Bautista, J. M., and Chen, N. (2020). Multiple intraspecific variations of mitochondrial genomes in the green-tide forming alga, Ulva compressa Linnaeus (Ulvophyceae, Chlorophyta). Front. Mar. Sci. 7:714. doi: 10.3389/fmars.2020.00714
Liu, F., Pang, S., Chopin, T., Gao, S., Shan, T., Zhao, X., et al. (2013). Understanding the recurrent large-scale green tide in the Yellow Sea: temporal and spatial correlations between multiple geographical, aquacultural and biological factors. Mar. Environ. Res. 83, 38–47. doi: 10.1016/j.marenvres.2012.10.007
Liu, F., Pang, S., Chopin, T., Xu, N., Shan, T., Gao, S., et al. (2010). The dominant Ulva strain of the 2008 green algal bloom in the Yellow Sea was not detected in the coastal waters of Qingdao in the following winter. J. Appl. Phycol. 22, 531–540. doi: 10.1007/s10811-009-9489-7
Liu, F., Zhang, Y., Bi, Y., Chen, W., and Moejes, F. W. (2019). Understanding the evolution of mitochondrial genomes in Phaeophyceae inferred from mitogenomes of Ishige okamurae (Ishigeales) and Dictyopteris divaricata (Dictyotales). J. Mol. Evol. 87, 16–26. doi: 10.1007/s00239-018-9881-5
Liu, J., Yang, X., Cui, J., Zhuang, M., Zhao, L., Li, J., et al. (2020). Complete chloroplast genome of Ulva meridionalis (Ulvales: Ulvaceae): an extremely fast-growing green macroalgae. Mitochondrial DNA Part B 5, 1390–1392. doi: 10.1080/23802359.2020.1735967
Luo, R., Liu, B., Xie, Y., Li, Z., Huang, W., Yuan, J., et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18.
Marshall, K., Joint, I., Callow, M. E., and Callow, J. A. (2006). Effect of marine bacterial isolates on the growth and morphology of axenic plantlets of the green alga Ulva linza. Microb. Ecol. 52, 302–310. doi: 10.1007/s00248-006-9060-x
Melton, J. T., Leliaert, F., Tronholm, A., and Lopez-Bautista, J. M. (2015). The complete chloroplast and mitochondrial genomes of the green macroalga Ulva sp. UNA00071828 (Ulvophyceae, Chlorophyta). PLoS One 10:e0121020. doi: 10.1371/journal.pone.0121020
Melton, J. T., and Lopez-Bautista, J. M. (2017). The chloroplast genome of the marine green macroalga Ulva fasciata Delile (Ulvophyceae, Chlorophyta). Mitochondrial DNA Part A 28, 93–95. doi: 10.3109/19401736.2015.1110814
Mine, I., Menzel, D., and Okuda, K. (2008). Morphogenesis in giant-celled algae. Int. Rev. Cell Mol. Biol. 266, 37–83. doi: 10.1016/s1937-6448(07)66002-x
Novikova, O., and Belfort, M. (2017). Mobile group II introns as ancestral eukaryotic elements. Trends Genet. 33, 773–783. doi: 10.1016/j.tig.2017.07.009
Pombert, J.-F., Lemieux, C., and Turmel, M. (2006). The complete chloroplast DNA sequence of the green alga Oltmannsiellopsis viridis reveals a distinctive quadripartite architecture in the chloroplast genome of early diverging ulvophytes. BMC Biol. 4:3. doi: 10.1186/1741-7007-4-3
Pombert, J.-F., Otis, C., Lemieux, C., and Turmel, M. (2005). The chloroplast genome sequence of the green alga Pseudendoclonium akinetum (Ulvophyceae) reveals unusual structural features and new insights into the branching order of chlorophyte lineages. Mol. Biol. Evol. 22, 1903–1918. doi: 10.1093/molbev/msi182
Punta, M., Coggill, P. C., and Eberhardt, R. Y. (2012). The Pfam protein families database. Nucleic Acids Res. 40, D290–D301.
Ruck, E. C., Nakov, T., Jansen, R. K., Theriot, E. C., and Alverson, A. J. (2014). Serial gene losses and foreign DNA underlie size and sequence variation in the plastid genomes of diatoms. Genome Biol. Evol. 6, 644–654. doi: 10.1093/gbe/evu039
Schattner, P., Brooks, A. N., and Lowe, T. M. (2005). The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33, 686–689.
Seetharaman, M., Eldho, N. V., Padgett, R. A., and Dayie, K. T. (2006). Structure of a self-splicing group II intron catalytic effect or domain 5: parallels with spliceosomal U6 RNA. RNA 12, 235–247. doi: 10.1261/rna.2237806
Sharma, M., Ellis, R. L., and Hinton, D. M. (1992). Identification of a family of bacteriophage T4 genes encoding proteins similar to those present in group I introns of fungi and phage. Proc. Natl. Acad. Sci. U. S. A. 89, 6658–6662. doi: 10.1073/pnas.89.14.6658
Spoerner, M., Wichard, T., Bachhuber, T., Stratmann, J., and Oertel, W. (2012). Growth and thallus morphogenesis of Ulva mutabilis (Chlorophyta) depends on a combination of two bacterial species excreting regulatory factors. J. Phycol. 48, 1433–1447. doi: 10.1111/j.1529-8817.2012.01231.x
Steinhagen, S., Barco, A., Wichard, T., and Weinberger, F. (2019a). Conspecificity of the model organism Ulva mutabilis and Ulva compressa (Ulvophyceae, Chlorophyta). J. Phycol. 55, 25–36. doi: 10.1111/jpy.12804
Steinhagen, S., Weinberger, F., and Karez, R. (2019b). Molecular analysis of Ulva compressa (Chlorophyta, Ulvales) reveals its morphological plasticity, distribution and potential invasiveness on German North Sea and Baltic Sea coasts. Eur. J. Phycol. 54, 102–114. doi: 10.1080/09670262.2018.1513167
Stoddard, B. L. (2011). Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification. Structure 19, 7–15. doi: 10.1016/j.str.2010.12.003
Straub, S. C., Cronn, R. C., Edwards, C., Fishbein, M., and Liston, A. (2013). Horizontal transfer of DNA from the mitochondrial to the plastid genome and its subsequent evolution in milkweeds (apocynaceae). Genome Biol. Evol. 5, 1872–1885. doi: 10.1093/gbe/evt140
Suzuki, S., Yamaguchi, H., Hiraoka, M., and Kawachi, M. (2018). Mitochondrial and chloroplast genome sequences of Ulva ohnoi, a green-tide forming macroalga in the Southern coastal regions of Japan. Mitochondrial DNA Part B. 3, 765–767. doi: 10.1080/23802359.2018.1483778
Szitenberg, A., Rot, C., Ilan, M., and Huchon, D. (2010). Diversity of sponge mitochondrial introns revealed by cox 1 sequences of Tetillidae. BMC Evol. Biol. 10:288. doi: 10.1186/1471-2148-10-288
Tamura, K., and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526.
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The ClustalX windows interface flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tritt, A., Eisen, J. A., Facciotti, M. T., and Darling, A. E. (2012). An integrated pipeline for de novo assembly of microbial genomes. PLoS One 7:e42304. doi: 10.1371/journal.pone.0042304
Turmel, M., and Lemieux, C. (2018). Evolution of the plastid genome in green algae. Adv. Bot. Res. 85, 157–193. doi: 10.1016/bs.abr.2017.11.010
Turmel, M., Otis, C., and Lemieux, C. (2016). Mitochondrion-to-chloroplast DNA transfers and intragenomic proliferation of chloroplast group II introns in Gloeotilopsis green algae (Ulotrichales, Ulvophyceae). Genome Biol. Evol. 8, 2789–2805. doi: 10.1093/gbe/evw190
Turmel, M., Otis, C., and Lemieux, C. (2017). Divergent copies of the large inverted repeat in the chloroplast genomes of ulvophycean green algae. Sci. Rep. 7:994.
VanRoey, P., Meehan, L., Kowalski, J. C., Belfort, M., and Derbyshire, V. (2002). Catalytic domain structure and hypothesis for function of GIY-YIG intron endonuclease I-TevI. Nat. Struct. Biol. 9, 806–811.
Wang, L., Cai, C., Zhou, L., He, P., and Jiao, B. (2017). The complete chloroplast genome sequence of Ulva linza. Conserv. Genet. Resour. 9, 463–466. doi: 10.1007/s12686-016-0682-0
Wichard, T., Charrier, B., Mineur, F., Bothwell, J. H., De Clerck, O., and Coates, J. C. (2015). The green seaweed Ulva: a model system to study morphogenesis. Front. Plant Sci. 6:72. doi: 10.3389/fpls.2015.00072
Wyman, S. K., Jansen, R. K., and Boore, J. L. (2004). Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20, 3252–3255. doi: 10.1093/bioinformatics/bth352
Zhu, H., Hu, Y., Liu, F., Hu, Z., and Liu, G. (2019). Characterization of the chloroplast genome of Trentepohlia odorata (Trentepohliales, Chlorophyta), and discussion of its taxonomy. Int. J. Mol. Sci. 20:1774. doi: 10.3390/ijms20071774
Keywords: chloroplast genome, green tide, Ulvophyceae, green algae, group I/II intron, intron-encoded protein
Citation: Liu F and Melton JT III (2021) Chloroplast Genomes of the Green-Tide Forming Alga Ulva compressa: Comparative Chloroplast Genomics in the Genus Ulva (Ulvophyceae, Chlorophyta). Front. Mar. Sci. 8:668542. doi: 10.3389/fmars.2021.668542
Received: 16 February 2021; Accepted: 24 March 2021;
Published: 15 April 2021.
Edited by:
Andrew Stanley Mount, Clemson University, United StatesReviewed by:
Georg Hausner, University of Manitoba, CanadaYongyu Zhang, Qingdao Institute of Bioenergy and Bioprocess Technology (CAS), China
Copyright © 2021 Liu and Melton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Liu, liufeng@qdio.ac.cn; prcliufeng@sina.cn
 Feng Liu
Feng Liu James T. Melton III
James T. Melton III