First Record of Bioluminescence in a Sipunculan Worm
- 1Departamento de Oceanografia Física, Instituto Oceanográfico, Química e Geológica, Universidade de São Paulo, São Paulo, Brazil
- 2Departamento de Biologia, Centro de Ciências Humanas e Biológicas, Universidade Federal de São Carlos, Sorocaba, Brazil
- 3Department of Marine Biology, Texas A&M University at Galveston, Galveston, TX, United States
During a search for bioluminescent marine annelids around the island of Carrie Bow Cay, Belize, we discovered bioluminescence in the sipunculan worm Nephasoma pellucidum. The identification of N. pellucidum was verified by DNA barcoding of ∼650 bp of the mitochondrial cytochrome c oxidase I gene, using sequences of the same species from previous studies as references. Our study constitutes the first record of a bioluminescent sipunculan species. Green luminescence was detected in four of the five individuals of N. pellucidum collected from coral rubble from a shallow reef flat and one deeper (14 m) site. To test for bioluminescent activity, all collected annelids (including sipunculans) were sorted into morphotypes and identified to the lowest taxonomic level possible. After several hours of dark adaptation, they were mechanically stimulated, and luminescent reactions were noted and recorded by photography and/or video whenever possible. Of a total of nine examined sipunculan species, N. pellucidum was the only one that showed bioluminescent activity. The underlying biochemical processes for the bioluminescence are so far unknown, but transcriptome data indicate the presence of proteins with similarities to Renilla-type luciferases. We did not find similarities to any known photoproteins. Our findings broaden the known diversity of bioluminescent annelid taxa and support the notion that bioluminescence arose multiple times in annelids. The bioluminescent properties of N. pellucidum add a new reason to pursue this species as a model species, as has been previously proposed.
Introduction
Bioluminescence, the biochemical generation of cold, visible light, is a very common phenomenon in marine organisms, especially in the deep sea (Haddock et al., 2010; Widder, 2010). Lau and Oakley (2021) estimate that bioluminescence has evolved 94 times across the tree of life. In animals, bioluminescence has been reported in at least 12 phyla and often appears to have multiple evolutionary origins within a given phylum (Haddock et al., 2010; Waldenmaier et al., 2012; Lau and Oakley, 2021). While the bioluminescent properties of some phyla, especially cnidarians and arthropods, have been studied extensively, comparatively little is known about bioluminescence in the phylum Annelida. Annelids include at least 13 family-level taxa with reported bioluminescent representatives, as well as several additional anecdotal reports (Verdes and Gruber, 2017). Annelid bioluminescence spans terrestrial taxa and diverse marine groups. There is no single habitat in which marine annelids are particularly prone to displaying bioluminescence nor a particular body region that produces light. Some examples of bioluminescent annelids include the tube-dwelling chaetopterids (Nicol, 1952b, c, a, 1957; Mirza et al., 2020), epifaunal taxa such as polynoid scale worms (Nicol, 1954; Herrera, 1979; Plyuscheva and Martin, 2009; Moraes et al., 2021), and pelagic forms such as tomopterids (Dales, 1971; Gouveneaux et al., 2017). Additionally, there is a wide variety of wavelengths of emitted light, from yellow to blue (Verdes and Gruber, 2017). Ecological functions of bioluminescence include intraspecific communication, especially mate attraction (e.g., Galloway and Welch, 1911; Fischer and Fischer, 1995) and predation deterrence (e.g., Livermore et al., 2018) but in many cases the ecological roles of annelid bioluminescence are not well understood.
The classical biochemical pathway for the generation of light involves a small molecule, generically called luciferin, and an enzyme, known as luciferase, which catalyzes the oxidation of luciferin to oxyluciferin. This reaction produces an unstable complex (usually a cyclic peroxide) that breaks down to produce a compound called oxyluciferin and gives off a large amount of energy as light (Shimomura, 2006). Some luciferases form complexes with the substrate, known as photoproteins, which may also depend on cofactors such as Ca2+, Fe2+, H2O2, and ATP, among others (Daunert and Deo, 2006). In annelids, the best studied bioluminescent pathway is that of Odontosyllis. Odontosyllis is a genus of syllid polychetes inhabiting shallow coastal warm-water habitats. Benthic as adults, they periodically produce bioluminescent swimming stages that rise to the surface and release likewise bioluminescent gametes. The bioluminescent properties are retained post-spawning, suggesting that the ecological function of bioluminescence is not limited to reproduction (Fischer and Fischer, 1995). This green/blue bioluminescent system has been identified as a unique luciferin/luciferase system which requires no cofactor and shows little resemblance to those of other organisms (Brugler et al., 2018; Schultz et al., 2018; Kotlobay et al., 2019).
Here we present the first report of a bioluminescent sipunculan worm. Sipunculan worms, also known as peanut worms or star worms, represent an understudied group of unsegmented marine worms, characterized by a simple sac-like body and a retractable introvert with a crown of tentacles at its tip (Cutler, 1994; Schulze et al., 2019; Schulze and Kawauchi, 2021). Sipuncula were long regarded as a distinct phylum but recent phylogenetic and phylogenomic studies (Struck et al., 2007; Mwinyi et al., 2009; Dordel et al., 2010; Weigert et al., 2014), have placed them as a basal branch in the annelid radiation, despite their lack of segmentation (Carrillo-Baltodano et al., 2019). Sipunculans generally have a cryptic lifestyle and are rarely seen unless actively sought after. One habitat where sipunculans are particularly abundant and diverse is coral rubble, or eroded pieces of coral skeleton where sipunculans either borrow into the calcareous material or inhabit existing holes and crevices (Rice, 1975; Rice and MacIntyre, 1982; Gómez et al., 2013). Many of these species have long introverts, with recurved hooks that they likely use to scrape epiphytic organisms from surfaces (Rice, 1975, 1976; Cutler, 1994), but otherwise little is known about their biology.
The discovery of bioluminescence in a single sipunculan species from Carrie Bow Cay (CBC), Belize was the result of a search for novel bioluminescent annelid systems focusing on shallow-water tropical taxa. Although the region around CBC has been thoroughly sampled for sipunculans during previous expeditions (Rice and MacIntyre, 1982; Schulze and Rice, 2004), the bioluminescent species we observed, Nephasoma pellucidum (Keferstein, 1865), has not previously been reported from CBC. However, it has been reported from more southern Caribbean locations, such as Bocas del Toro, Panama (Schulze, 2005), Curaçao and Venezuela (Dean et al., 2007), as well as in the western Atlantic (Schulze and Rice, 2009), eastern Pacific (Dean and Cutler, 1998) and occasionally from the Indo-Pacific (Cutler, 1994; Kawauchi et al., 2012). Although it is not a particularly abundant species in any of the known Caribbean/Atlantic locations, N. pellucidum has been proposed as a model species for spiralian development (Schulze and Rice, 2009; Boyle and Rice, 2014), because it is relatively easy to keep in laboratory culture. Males and females from the Florida population readily spawn in the lab and larval development is well documented (Schulze and Rice, 2009), although attempts to complete the life cycle through metamorphosis have so far been unsuccessful. Here, we report on our methods of collection and eliciting the bioluminescent response.
Materials and Methods
Collections and Morphological Identification
Field collections were conducted at the Smithsonian CBC Field Station in February of 2020. CBC is a small island located at 16.8000°N, 88.0833°W, about 24 kilometers off the coast of Belize, on the southern portion of the Meso-American Barrier Reef. The east side of the island is flanked by a reef flat, followed by the reef crest and a drop-off to deeper waters. The landward (west) side of the island is dominated by seagrass and patch reefs. Coral reef, seagrass, and intertidal coral rubble habitats were all sampled following a similar protocol. Pieces of coral rubble were collected in mesh dive bags or buckets either by SCUBA, snorkel, or wading into the water. The collected material was kept in free-flowing saltwater tables until processing. Pieces of coral rubble were carefully cracked using a hammer and chisel. Sipunculans were found either embedded in the calcium carbonate skeletons or nestled in grooves within the rubble. Specimens were extracted using forceps and blunt probes.
All specimens were then kept in small plastic dishes covered with seawater which was periodically replaced to prevent fouling. Individuals were sorted into morphotypes and identified to the lowest taxonomic level possible. Multiple individuals of the same identification were pooled. All dishes were placed underneath a dark box. Specimens were left in this configuration for the dark acclimation period prior to bioluminescence screening. The identification of the bioluminescent species was confirmed after return from CBC by a closer inspection of the morphology, dissections to observe the internal anatomy, and through molecular taxonomy (DNA barcoding, see section “Molecular Taxonomy”).
Molecular Taxonomy
We used DNA the barcoding sequence of the cytochrome c oxidase subunit I gene (COI) (Hebert et al., 2003) to confirm the morphology-based identification. DNA was extracted from small pieces of the introvert retractor muscles and body wall from the two injured specimens. DNA extraction was performed using the Qiagen DNEasy Blood and Tissue kit, following the instructions of the manufacturer. The COI barcoding sequence was amplified using the primer combination Poly-LCO (GAYTATWTTCAACAAATCATAAAGATATTGG) and Poly-HCO (TAMACTTCWGGGTGACCAAARAATCA) (Carr et al., 2011). The PCR mix consisted of 12.5 μl OneTaq Mastermix (New England Biolabs), 0.5 μl magnesium chloride (25 mM), 1 μl of each primer (10 μM), 9 μl water, and 1 μl of genomic DNA template. The cycling protocol was as follows: 94°C for 30 s, 35 cycles of 94°C for 30 s, 42°C for 30 s, and 68°C for 60 s; followed by a final extension at 68°C for 5 min. Successful amplification was confirmed through electrophoresis on 1% agarose gels. Amplification was only successful for one of the two specimens. The PCR product was enzymatically cleaned using the ExoSapIt Express reagent (ThermoFisher) and sequenced in both directions at Genewiz1. The forward and reverse fragments were assembled in CodonCode Aligner v9.0.22. The single sequence with a length of 658 bp obtained from one specimen is available at GenBank under accession number MZ852770. A BLAST search for this sequence in GenBank resulted in multiple close hits, therefore we performed a phylogenetic analysis to determine the specimen’s closest relatives. The obtained sequence was aligned with publicly available COI sequences from other members of the genera Nephasoma, including two identified as N. pellucidum, and Golfingia, the presumed sister taxon to Nephasoma, as well as one sequence for Siphonosoma cumanense as an outgroup. Alignment was performed using Muscle algorithm in Geneious Prime® 2021.2.2. The final alignment was 649 bp long. Phylogenetic analysis was conducted using Bayesian Inference in MrBayes on XSEDE through the CIPRES Science Gateway (Miller et al., 2010). Bayesian analysis was performed under a GTR+I+G model. Two runs with four chains each were run for 10,000,000 generations of MCMC, sampling every 1,000th tree. A 50% consensus tree was calculated after discarding trees from the first 2,500,000 generations.
Observation and Documentation of Bioluminescence
Specimens were acclimated to the dark for at least 5 h. The dark acclimation effectively eliminates any potential stress from an increase in non-directional light intensity caused by the extraction out of their concealed burrows (embedded in rock, or rubble). We retrofitted the field station’s laboratory into a functional darkroom by covering all windows and light sources. We then spent 15–20 min acclimating our eyes to the darkness. At this point, specimens were removed from the dark box and screened for bioluminescence by mechanically irritating them with blunt probes or forceps for 5–10 s. Any sighting of visible light in response to this stimulation was recorded as a positive display of bioluminescence.
Additional individuals of positive bioluminescent species were then targeted in field collections and similarly screened. Bioluminescent activity was documented using a Sony α7 s digital camera attached to a Sony SEL50M28 lens, with an ISO setting of 32,000 and a wide aperture (f/2.8), producing a video of 24 frames per second. We used red light, which does not seem to interfere with light production, to position the specimens for video and photo documentation. Post-processing of video contrast and exposure were enhanced for visualization in the video editor Adobe Premiere Pro 2019. Individuals with confirmed bioluminescent displays were preserved in either 95% ethanol or RNAlater for genetic analysis, or 10% formaldehyde for taxonomic purposes.
Transcriptome Analysis
The transcriptome raw datasets already sequenced and available in a public database for nine Sipuncula species (Andrade et al., 2015; Lemer et al., 2015; Table 1), including one of N. pellucidum from Florida, were used for the prospection of genes associated with the bioluminescence in N. pellucidum. The read datasets were downloaded and filtered to remove low-quality reads and adaptors with SeqyClean v.1.10.09 (Zhbannikov et al., 2017) and used for de novo assembly in Trinity v.2.11.0 (Grabherr et al., 2011), with the default settings and in silico normalization. The transcripts with more than 200 bp were translated to amino acids by TransDecoder v.5.5.0 and were subjected to a similarity search against the non-redundant (nr) SWISS-PROT v.5 database using the BLASTp (Altschul et al., 1997) algorithm, with a cut-off e-value of ≤10–5. We also conducted a second annotation step using specific gene products related to luciferase/photoprotein and GFP (green fluorescent protein) present in public databases. For that, we promoted a search in the non-redundant NCBI database using the terms “Luciferase,” “Photoprotein,” “Aequorin,” “Obelin,” “ Mnemiopsin,” “GFP,” and “Green Fluorescent Protein” (Access retrieve: October 5, 2021) and created a local database using makeblastdb function. The transcripts abundance was calculated by the align_and_estimate_abundance.pl script, which is present in the Trinity package. We estimated the completeness of the transcriptome against the metazoa database using BUSCO v.4.0.1 (Simão et al., 2015) software. The transcriptomes assembly and the photoprotein/luciferase sequences databases were deposited in the FigShare repository (10.6084/m9.figshare.16574831).
Results
Collection and Identification
In addition to N. pellucidum, we collected eight other sipunculan species from rubble around CBC: Aspidosiphon sp., Aspidosiphon cf. steenstrupii; Aspidosiphon cf. laevis; Aspidosiphon cf. elegans; Aspidosiphon cristatus, Phascolosoma nigrescens; Phascolosoma perlucens, and Antillesoma antillarum. Members of the genus Nephasoma have an array of tentacles arranged around the mouth at the tip of the introvert which is generally shorter than or approximately equal in length to the trunk. In addition, they have two introvert retractor muscles and two nephridia. Although the introverts in our preserved samples were retracted, we had observed the extended introverts with the tentacles in the live specimens (Visible in Supplementary Videos). The dissection of the preserved material showed the two retractor muscles and paired nephridia, confirming the genus identification. The two preserved specimens had trunk lengths of 34.4 and 45 mm, respectively (note that the length depends on the contraction status, Figures 1A,C). The trunk and proximal introvert of N. pellucidum are pale in color and covered by raised, often dark papillae. The two retractor muscles insert in the ventral body wall approximately 50% down the trunk. The nephridia are dark and not attached to the body wall. Nephridiopores are located at the level of the anus. Cutler (1994) mentions the presence of two dark eyespots on the brain which we observed as well. Over the course of 1 week, five specimens of N. pellucidum were recovered from coral rubble in shallow water (<2 m) around CBC and one specimen from coral rubble at Raph’s Wall (16.77961°N; 88.075117°W) from ∼14 m depth. Two of them were injured during retrieval from the rock, one of which was immediately fixed in 95% ethanol.
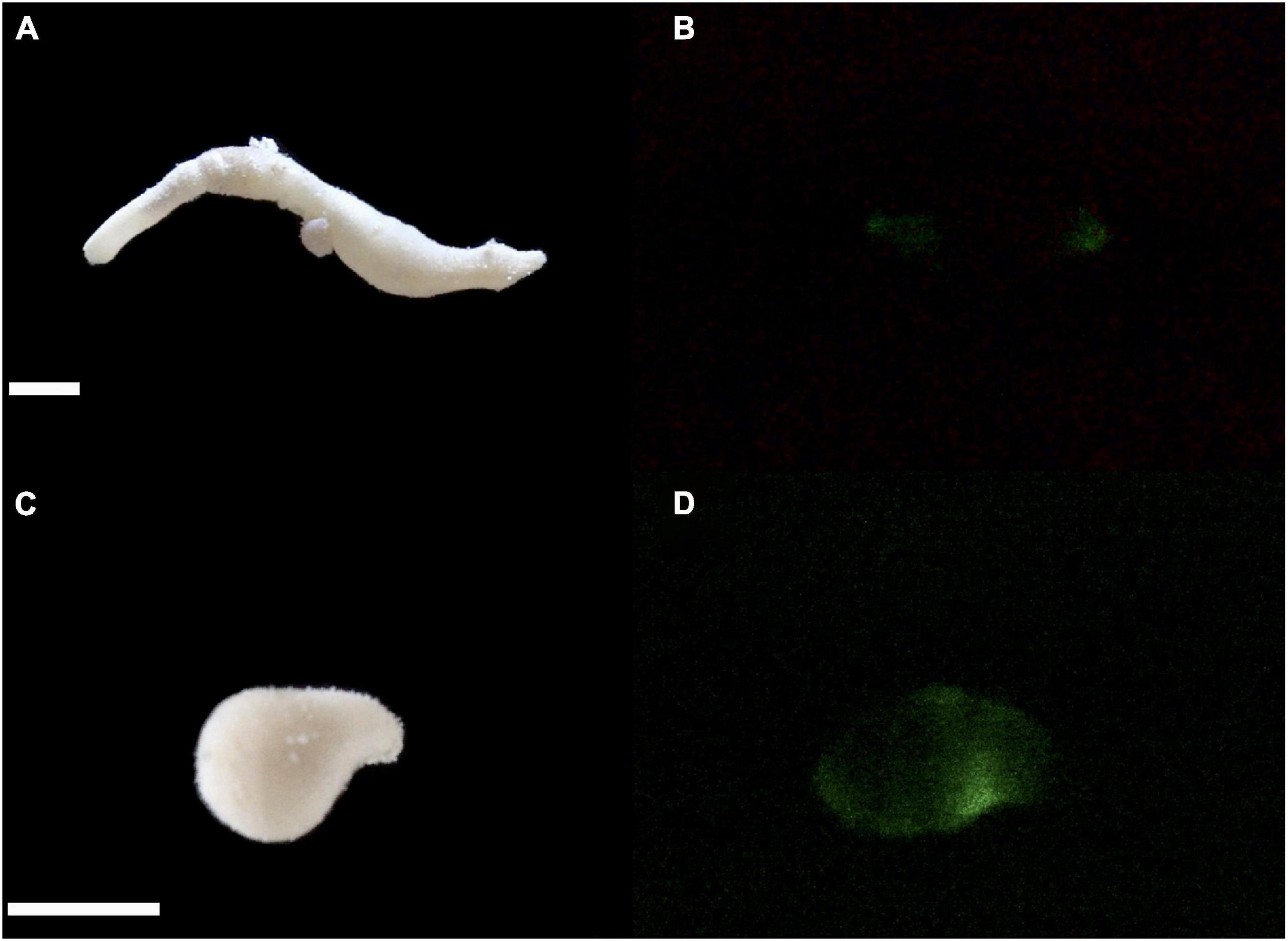
Figure 1. Images of N. pellucidum with introvert everted (A), retracted (C) and in the dark displaying bioluminescence by mechanical stimulation (B,D). Scale bars = 0.5 cm. Photos: Oliveira G. Anderson. Images were processed using Adobe Premiere Pro 2019.
Observed Bioluminescence
Out of the nine sipunculan species identified, only N. pellucidum produced light upon stimulation (Figure 1B,D). We observed bioluminescence in four different specimens, one from Raph’s wall and three from the shallow water rubble. The remaining fifth individual of N. pellucidum was not tested for bioluminescence due to injury upon extraction. Each specimen produced a pulse of visible greenish light upon perturbations with a blunt probe (Supplementary Video). The glow was localized on the body wall with no apparent patterning, although the intensity was not even throughout the body.
Molecular Taxonomy
The phylogenetic analysis placed N. pellucidum from Belize in a clade with N. pellucidum from New Caledonia and Florida, plus one N. flagriferum sequence (Figure 2). This clade is strongly supported by 100% posterior probability. Kimura-2-Parameter distances among the N. pellucidum sequences are: Belize-New Caledonia: 22.8%; Belize-Florida: 22.4%; and Florida-New Caledonia: 21.9%. Sequence variation is almost entirely in the third codon position, with only a single amino acid difference between the Florida sequence and the other two.
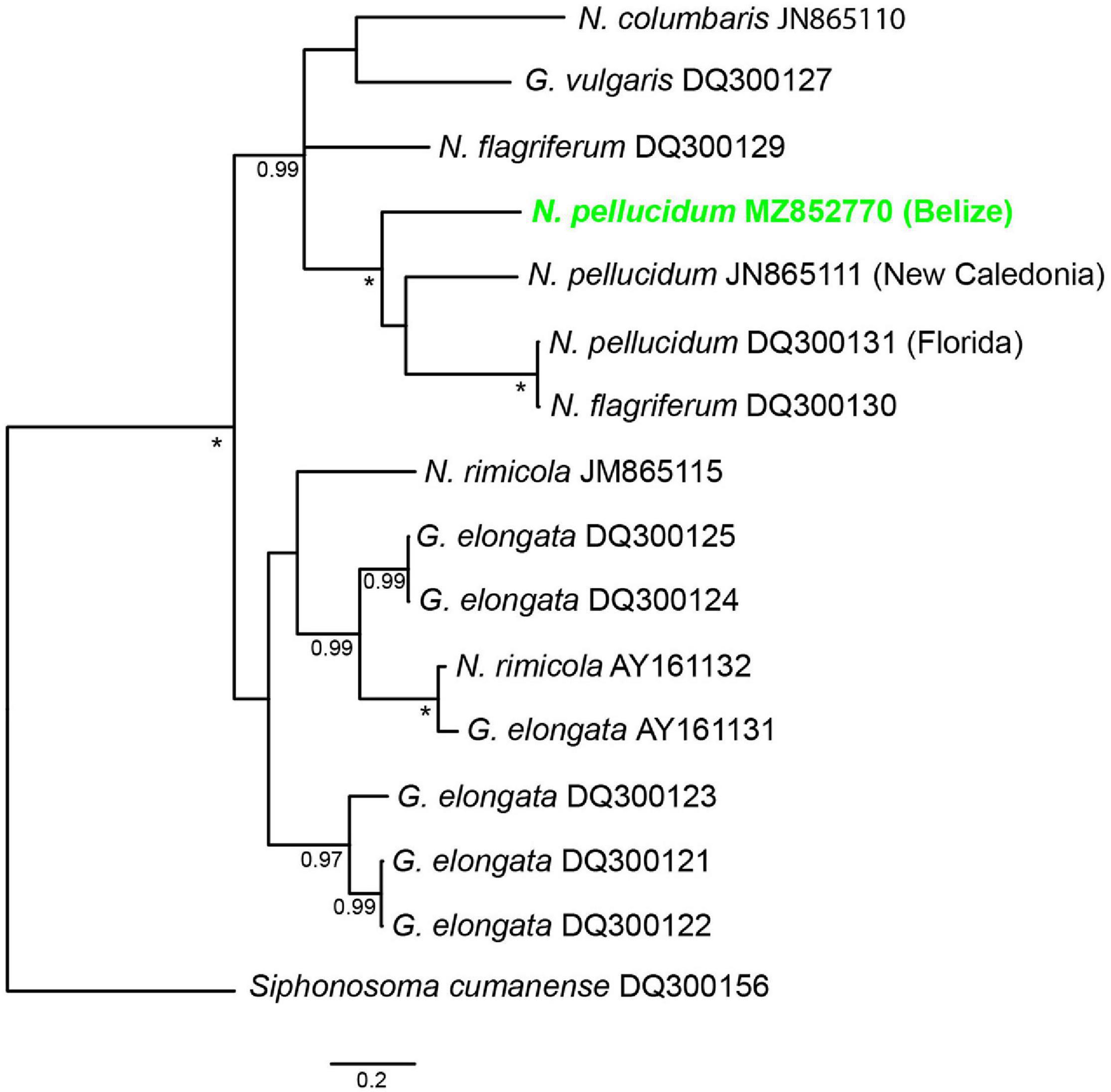
Figure 2. 50% majority rule consensus tree resulting from Bayesian analysis of available COI sequences for Nephasoma and Golfingia species. Posterior probabilities over 90% are indicated at the notes; asterisks (*) indicate 100% posterior probability. All sequences are labeled with the species name and Genebank accession code.
Transcriptomic Findings
A total of 115,867 gene products were obtained for N. pellucidum, which correspond to 39.22% of transcriptome completeness. After the amino acid translation filtering, we obtained a total of 13,815 coding sequences, in which 7,172 (∼52%) were annotated. Among them, we identified gene products associated with the bioluminescence in Renilla, echinoderms, starfish, and firefly, which are related to luciferase, coelenterazine, and Renilla luciferin/luciferase. We have also carried out the second round of annotation using a constrain search against luciferases, from metazoa to bacteria (identified as responsible for luminescent emission in some marine species, Delroisse et al., 2021), and Fluorescent Proteins FPs such as GFPs (green fluorescent proteins, Rodriguez et al., 2017), which may explain the greenish emission in this species. We observed gene product similarity to coelenterazine 2-monooxygenase, Renilla-luciferin monooxygenase, Renilla luciferase, firefly luciferase/4-Coumarate CoA-ligase, and firefly luciferin sulfotransferase present in bioluminescent (eg., Photinus pyralis and Luciola cruciata) and non-bioluminescent species (eg., Strongylocentrotus purpuratus and Lytechinus variegatus; Supplementary Table 1), however, no similarity was observed for other photoproteins and green fluorescent proteins. The evaluation of transcript abundance, in FPKM (Fragment per Kilobase per Million mapped bases), showed lower values for all the transcripts identified (Supplementary Table 1).
We also carried out the assembly and annotation of the RNA-Seq available from the other 8 species of Sipuncula for comparative purposes (details about the assembly results available in Supplementary Table 1). As described for N. pellucidum, no similarity was observed to photoproteins, green fluorescent proteins, or bacterial luciferase. However, we have found gene products related to luciferase and Renilla luciferin/luciferase in five of them (Sipunculus nudus, A. antillarum, Aspidosiphon parvulus, Golfingia vulgaris, and Phacolopsis gouldii). The other three species (Phascolion cryptum, P. perlucens, and S. cumanense) displayed a low number of coding sequences, which may explain why we did not recover any of these gene products.
Discussion
Implications for the Diversity of Bioluminescent Systems in Annelids
The unexpected discovery of bioluminescence in Sipuncula highlights the current lack of knowledge about the distribution, evolution, ecological functions, and mechanisms of bioluminescence in annelids. Although we did not observe bioluminescence in any of the other sipunculan species we collected, sipunculans should be considered in future exploratory studies of marine invertebrate bioluminescence. N. pellucidum is a derived species within the Sipuncula, belonging to a larger clade (referred to as Clade III) with poorly resolved taxonomy (Kawauchi et al., 2012; Lemer et al., 2015; Schulze et al., 2007, 2019). None of the other species we collected in Belize fall into this clade; therefore, Clade III may be of particular interest. For many sipunculan species, the COI barcode sequences can be highly divergent within morphospecies, often leading to the detection of cryptic species (Schulze and Kawauchi, 2021). Based on the morphological and anatomical features of the specimens we collected, we are confident with the identification as N. pellucidum, despite the relatively high genetic distances to other specimens identified as such. The single sequence for Nephasoma flagriferum that falls into the same clade is suspiciously similar (a single nucleotide difference) to the Florida specimen of N. pellucidum and a mistake or sample contamination cannot be excluded for this sample. The transcriptome data analyzed here originated from an N. pellucidum specimen from Florida (Lemer et al., 2015). It is unknown whether this specimen had the same bioluminescent properties as our samples from Belize. Future studies on N. pellucidum specimens from other locations, combined with transcriptomic analysis from confirmed luminescing specimens, will therefore be important.
Assuming that bioluminescence evolved multiple times independently in annelids, it is difficult to make parallels between the few mechanisms already discovered (Haddock et al., 2010; Verdes and Gruber, 2017). For example, the bioluminescent system of the tubeworm Chaetopterus variopedatus has been studied since the early 1960s (Shimomura, 2006). Shimomura and Johnson isolated and crystallized a photoprotein that emitted light in the presence of Fe2+, peroxides and molecular oxygen (Shimomura and Johnson, 1968). Additionally, two other unknown activating factors also stimulated light emission in vitro. However, knowledge about the bioluminescence of the C. variopedatus is still fragmented and incomplete, due mainly to the numerous difficulties related to the isolation of the luminous components involved in this process (Mirza et al., 2020).
Regarding the Polynoidae family, Nicolas et al. (1982) isolated and partially purified a photoprotein that would be responsible for the light emission in the scale worm Harmothoe lunulata. According to some authors, the homogenate prepared from the luminous scales was capable of emitting light through the addition of various reagents, such as sodium dithionite (Herrera et al., 1974), ferrous ions (Lecuyer and Arrio, 1975), xanthine-xanthine oxidase system, and Fenton reaction (Nicolas et al., 1979). The isolated 66 kDa photoprotein was named polynoidin, but there is no additional information available on the chemical studies of bioluminescence in Harmothoe, nor any proposal for a chemical structure for its luciferin or methodologies described to isolate it (Nicolas et al., 1982; Shimomura, 2006; Moraes et al., 2021).
Recently, the isolation and structural characterization of a novel luciferin from the fireworm Odontosyllis (Syllidae) was published by Kotlobay et al. (2019). By using the polychete Odontosyllis undecimdonta as a source of luminogenic material, the authors showed that an unusual tricyclic sulfur-containing heterocycle serves as a substrate in the light emission by this syllid (Kotlobay et al., 2019). Also, Mitani et al. (2019) demonstrated that the luciferase gene of O. undecimdonta has a different evolutionarily origin, since this gene has no homology to any other known sequence from a marine bioluminescent system.
The evaluation of transcriptome analyses may suggest three hypotheses to explain the bioluminescence in N. pellucidum: (i) a possible association among the enzymes related to firefly and/or Renilla luciferase to the bioluminescence emission, (ii) the bioluminescence is caused by distinct sources, such as symbiotic bacteria, although we did not recover any gene product associated with the bacterial luciferase, or (iii) the presence of an unknown (and new) bioluminescent system.
Regarding hypothesis (i), our results obtained from the N. pellucidum transcriptome suggest the presence of gene products associated with firefly ligases and sulfotransferases. However, it appears unlikely that any firefly luminescent protein is involved in the light emission mechanism of N. pellucidum, since the biochemistry system (including an unknown luciferin) and the photophore morphologic structure are quite distinct between these species. Additionally, other genes related to bioluminescent products from the luminous sea pansy Renilla reniformis, such as a Renilla-type luciferase, were also found. Considering the green bioluminescence of the N. pellucidum species and the low expression of the Renilla-type luciferase candidates it is important to clearly state that while these Renilla-type luciferases will be interesting to investigate in the future, there is no indication of the potential involvement of these enzymes in the bioluminescence of the species.
The luciferase present in the soft coral Renilla is responsible for catalyzing the oxidation of the coelenterazine molecule, a luciferin found in at least eight different aquatic phyla, producing blue light (Jiang et al., 2016; Vacher et al., 2018). Whilst, a Renilla-type luciferase presents great tertiary structure similarity, as well as homology, with bacterial haloalkane dehalogenases (Loening et al., 2006; Delroisse et al., 2017). Recently, Renilla-type luciferases have been identified in the brittle star Amphiura filiformis (Echinodermata, Ophiuroidea) (Delroisse et al., 2017) as well as in the tunicate Pyrosoma atlanticum (Chordata, Thaliacea) (Tessler et al., 2020). The bioluminescence color attributed to Renilla-type luciferases present in A. filiformis and P. atlanticum is blue, which does not explain the green color produced by N. pellucidum.
In corals such as Renilla, the blue light generated by the oxidation of coelenterazine is modulated to a green color through a GFP (Green Fluorescent Protein), through a process of resonance energy transfer (Shimomura, 2006). However, no gene homologous to a GFP or any other fluorescent protein was found in the N. pellucidum transcriptome. Yet, some coelenterazine-dependent photoproteins present in a few species of ctenophores are capable of emitting light at wavelengths shifted to the color green, but again no homologous photoprotein genes were detected (Haddock and Case, 1999). Future biochemical experiments will be necessary to corroborate a potential role, or not, of coelenterazine in the bioluminescence of N. pellucidum. In addition, in the case of a new bioluminescent system, the development of in vitro light emission assays will be necessary to isolate and purify the luminogenic molecular components of this system.
Considering the great diversity of bioluminescence mechanisms in marine annelids, the light emission process in N. pellucidum might also involve a new luminescent system (hypothesis ii). A final possibility (hypothesis iii) to be explored is that the bioluminescence in sipunculans is produced by bioluminescent bacteria, i.e., extrinsic bioluminescence. In fact, strains of marine bacteria of the Shewanella genus, known to contain bioluminescent species, have been isolated from sipunculans, such as Phascolosoma japonicum (Ivanova et al., 2003). For example, Shewanella baltica is a bioluminescent bacterium capable of emitting blue-greenish light (Burtseva et al., 2020). Evidently, all the hypotheses presented here require further investigation.
Conclusions and Future Research Directions
We have now reported on a novel bioluminescent system from a basal branch within the annelid radiation. This discovery has implications for further understanding the evolution, be it convergent or not, of this trait within Annelida. Future research directions include isolating and purifying the luminogenic substances and describing their properties, as well as exploring possible ecological functions. As for our transcriptome analysis, we now have targeted species, containing the luciferase-like gene products, for collection and bioluminescent screening. Additionally, as N. pellucidum can be maintained in laboratory culture, there is potential for studying the development of such trait or determining if the light production is diet derived.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MZ852770. The transcriptomes assembly and the photoprotein/luciferase sequences databases are available in FigShare (doi: 10.6084/m9.figshare.16574831).
Author Contributions
AO and AS conceived the project. AO, AS, and MH conducted the field and laboratory work. DA performed the bioinformatics analysis. All authors contributed to the writing of the manuscript.
Funding
This research was funded under a TAMU-FAPESP collaborative grant to AS and AO (2018/22304-5), FAPESP (2017/22501-2, 2020/07600-7, and 2018/22204-5) to AO, and Coordination for the Improvement of Higher Education Personnel (CAPES/PNPD-001) to DA.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Scott Jones and Jerry and Sandy Alanko for their support during our stay at Carrie Bow Cay. This research represents Caribbean Coral Reef Ecosystem (CCRE) contribution #1057. Samples were collected and exported to the United States under Marine Scientific Research Permit #0012-20 from the Belize Fisheries Department.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.762706/full#supplementary-material
Footnotes
References
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Andrade, S. C., Novo, M., Kawauchi, G. Y., Worsaae, K., Pleijel, F., Giribet, G., et al. (2015). Articulating “Archiannelids”: phylogenomics and annelid relationships, with emphasis on meiofaunal taxa. Mol. Biol. Evol. 32, 2860–2875. doi: 10.1093/molbev/msv157
Boyle, M. J., and Rice, M. E. (2014). Sipuncula: an emerging model of spiralian development and evolution. Int. J. Dev. Biol. 58, 485–495.
Brugler, M. R., Aguado, M. T., Tessler, M., and Siddall, M. E. (2018). The transcriptome of the Bermuda fireworm Odontosyllis enopla (Annelida: Syllidae): a unique luciferase gene family and putative epitoky-related genes. PLoS One 13:e0200944. doi: 10.1371/journal.pone.0200944
Burtseva, O., Kublanovskaya, A., Baulina, O., Fedorenko, T., Lobakova, E., and Chekanov, K. (2020). The strains of bioluminescent bacteria isolated from the White Sea finfishes: genera Photobacterium, Aliivibrio, Vibrio, Shewanella, and first luminous Kosakonia. J. Photochem. Photobiol. B Biol. 208:111895. doi: 10.1016/j.jphotobiol.2020.111895
Carr, C. M., Hardy, S. M., Brown, T. M., Macdonald, T. A., and Hebert, P. D. N. (2011). A tri-oceanic perspective: DNA barcoding reveals geographic structure and cryptic diversity in Canadian polychaetes. PLoS One 6:e22232. doi: 10.1371/journal.pone.0022232
Carrillo-Baltodano, A. M., Boyle, M. J., Rice, M. E., and Meyer, N. P. (2019). Developmental architecture of the nervous system in Themiste lageniformis (Sipuncula): New evidence from confocal laser scanning microscopy and gene expression. J. Morphol. 280, 1628–1650. doi: 10.1002/jmor.21054
Cutler, E. B. (1994). The Sipuncula. Their Systematics, Biology and Evolution. Ithaca, NY: Cornell University Press.
Dales, R. P. (1971). Bioluminescence in pelagic polychaetes. J. Fisher. Res. Board Canada 28, 1487–1489. doi: 10.1139/f71-228
Daunert, S., and Deo, S. K. (2006). Photoproteins in Bioanalysis. Weinheim: WILEY-VCH Verlag GmbH & Co. KGaA.
Dean, H. K., and Cutler, E. B. (1998). Range extension of Nephasoma pellucidum and new records of Apionsoma (Edmondsius) pectinatum (Sipuncula) and Thalassema steinbecki (Echiura) from the Pacific of Costa Rica. Rev. Biol. Trop. 46, 279–280.
Dean, H. K., Hernández-Ávila, I., and Cutler, E. B. (2007). Sipunculans of the caribbean coast of venezuela and curacçao. Zootaxa 1431, 45–54.
Delroisse, J., Duchatelet, L., Flammang, P., and Mallefet, J. (2021). Leaving the dark side? Insights into the evolution of luciferases. Front. Mar. Sci. 8:673620. doi: 10.3389/fmars.2021.673620
Delroisse, J., Ullrich-Lüter, E., Blaue, S., Ortega-Martinez, O., Eeckhaut, I., Flammang, P., et al. (2017). A puzzling homology: a brittle star using a putative cnidarian-type luciferase for bioluminescence. Open Biol. 7:160300. doi: 10.1098/rsob.160300
Dordel, J., Fisse, F., Purschke, G., and Struck, T. H. (2010). Phylogenetic position of Sipuncula derived from multi-gene and phylogenomic data and its implication for the evolution of segmentation. J. Zool. Syst. Evol. Res. 48, 197–207. doi: 10.1111/j.1439-0469.2010.00567.x
Fischer, A., and Fischer, U. (1995). On the life-style and life-cycle of the luminescent polychaete Odontosyllis enopla (Annelida: Polychaeta). Invert. Biol. 114, 236–247. doi: 10.2307/3226878
Galloway, T., and Welch, P. (1911). Studies on a phosphorescent Bermudan Annelid, Odontosyllis enopla Verrill. Transact. Am. Micros. Soc. 30, 13–39. doi: 10.2307/3221049
Gómez, C., Ardila, N., and Sanjuan-Muñoz, A. (2013). Sipunculans associated with dead coral skeletons in the Santa Marta region of Colombia, south-western Caribbean. J. Mar. Biol. Associat. U. K. 93, 1785–1793. doi: 10.1017/S002531541300043X
Gouveneaux, A., Flood, P. R., and Mallefet, J. (2017). Unexpected diversity of bioluminescence in planktonic worms. Luminescence 32, 394–400. doi: 10.1002/bio.3192
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Haddock, S. H. D., and Case, J. F. (1999). Bioluminescence spectra of shallow and deep-sea gelatinous zooplankton: ctenophores, medusae and siphonophores. Mar. Biol. 133, 571–582. doi: 10.1007/s002270050497
Haddock, S. H. D., Moline, M. A., and Case, J. F. (2010). Bioluminescence in the Sea. Ann. Rev. Mar. Sci. 2, 443–493. doi: 10.1146/annurev-marine-120308-081028
Hebert, P. D. N., Ratnasingham, S., and DeWaard, J. R. (2003). Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B Biol. Sci. 270, S96–S99.
Herrera, A. A. (1979). Electrophysiology of bioluminescent excitable epithelial cells in a polynoid polychaete worm. J. Compar. Physiol. 129, 67–78. doi: 10.1007/BF00679913
Herrera, A. A., Hastings, J. W., and Morin, J. G. (1974). Bioluminescence in cell free extracts of the scale worm Harmothoe (Annelida, Polynoidae). Biol. Bull. 147, 480–481.
Ivanova, E. P., Nedashkovskaya, O. I., Zhukova, N. V., Nicolau, D. V., Christen, R., and Mikhailov, V. V. (2003). Shewanella waksmanii sp. nov., isolated from a sipunculla (Phascolosoma japonicum). Int. J. Syst. Evol. Microbiol. 53, 1471–1477. doi: 10.1099/ijs.0.02630-0
Jiang, T., Du, L., and Li, M. (2016). Lighting up bioluminescence with coelenterazine: strategies and applications. Photochem. Photobiol. Sci. 15, 466–480. doi: 10.1039/c5pp00456j
Kawauchi, G. Y., Sharma, P. P., and Giribet, G. (2012). Sipunculan phylogeny based on six genes, with a new classification and the descriptions of two new families. Zool. Scr. 41, 186–210. doi: 10.1111/j.1463-6409.2011.00507.x
Keferstein, W. M. D. (1865). Beiträge zur anatomischen und systematischen kenntinss der sipunculiden. Nachr. Königl. Gesellschaft Wissenschaften Georg Augusts Univ. Göttingen 1865, 189–209.
Kotlobay, A. A., Dubinnyi, M. A., Purtov, K. V., Guglya, E. B., Rodionova, N. S., Petushkov, V. N., et al. (2019). Bioluminescence chemistry of fireworm Odontosyllis. Proc. Natl. Acad. Sci. U.S.A. 116:18911. doi: 10.1073/pnas.1902095116
Lau, E. S., and Oakley, T. H. (2021). Multi-level convergence of complex traits and the evolution of bioluminescence. Biol. Rev. 96, 673–691. doi: 10.1111/brv.12672
Lecuyer, B., and Arrio, B. (1975). Some spectral characteristics of the light emitting system of the polynoid worms. Photochem. Photobiol. 22, 213–215. doi: 10.1111/j.1751-1097.1975.tb06738.x
Lemer, S., Kawauchi, G. Y., Andrade, S. C. S., González, V. L., Boyle, J., and Giribet, G. (2015). Re-evaluating the phylogeny of Sipuncula through transcriptomics. Mol. Phylog. Evol. 83, 174–183. doi: 10.1016/j.ympev.2014.10.019
Livermore, J., Perreault, T., and Rivers, T. (2018). Luminescent defensive behaviors of polynoid polychaete worms to natural predators. Mar. Biol. 165:149. doi: 10.1007/s00227-018-3403-2
Loening, A. M., Fenn, T. D., Wu, A. M., and Gambhir, S. S. (2006). Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng. Des. Sel. 19, 391–400. doi: 10.1093/protein/gzl023
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES science gateway for the inference of large phylogenetic trees,” in Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov. 2010, New Orleans, LA, 1–8.
Mirza, J. D., Migotto, ÁE., Yampolsky, I. V., de Moraes, G. V., Tsarkova, A. S., and Oliveira, A. G. (2020). Chaetopterus variopedatus bioluminescence: a review of light emission within a species complex. Photochem. Photobiol. 96, 768–778. doi: 10.1111/php.13221
Mitani, Y., Yasuno, R., Futahashi, R., Oakley, T. H., and Ohmiya, Y. (2019). Luciferase gene of a Caribbean fireworm (Syllidae) from Puerto Rico. Sci. Rep. 9:13015. doi: 10.1038/s41598-019-49538-7
Moraes, G. V., Hannon, M. C., Soares, D. M. M., Stevani, C. V., Schulze, A., and Oliveira, A. G. (2021). Bioluminescence in polynoid scale worms (Annelida: Polynoidae). Front. Mar. Sci. 8:643197. doi: 10.3389/fmars.2021.643197
Mwinyi, A., Meyer, A., Bleidorn, C., Lieb, B., Bartolomaeus, T., and Podsiadlowski, L. (2009). Mitochondrial genome sequence and gene order of Sipunculus nudus give additional support for an inclusion of Sipuncula into Annelida. BMC Genom. 10:27. doi: 10.1186/1471-2164-10-27
Nicol, J. A. C. (1952a). Studies on Chaetopterus variopedatus (Renier). I. The light-producing glands. J. Mar. Biol. Associat. U. K. 30, 417–433. doi: 10.1017/S002531540001290X
Nicol, J. A. C. (1952b). Studies on Chaetopterus variopedatus (Renier). II. Nervous control of light production. J. Mar. Biol. Associat. U. K. 30, 433–452. doi: 10.1017/S0025315400012911
Nicol, J. A. C. (1952c). Studies on Chaetopterus variopedatus (Renier). III. Factors affecting the light response. J. Mar. Biol. Associat. U. K. 31, 113–144. doi: 10.1017/S0025315400003738
Nicol, J. A. C. (1954). The nervous control of luminescent responses in polynoid worms. J. Mar. Biol. Associat. U. K. 33, 225–255. doi: 10.1017/S002531540000357X
Nicol, J. A. C. (1957). Spectral composition of the light of Chaetopterus. J. Mar. Biol. Associat. U. K. 36, 629–642. doi: 10.1017/S0025315400025893
Nicolas, M. T. (1979). Presence de photosomes dans les fractions lumineuses du système elytral des Polynoinae (Annelides, Polychetes). C. R. Acad. Sci. Paris 289, 177–180.
Nicolas, M. T., Bassot, J. M., and Shimomura, O. (1982). Polynoidin, a membrane photoprotein isolated from the bioluminescent system of scale- worms. Photochem. Photobiol. 35, 201–207. doi: 10.1111/j.1751-1097.1982.tb03832.x
Plyuscheva, M., and Martin, D. (2009). On the morphology of elytra as luminescent organs in scale-worms (Polychaeta, Polynoidae). Zoosymposia 2, 379–389.
Rice, M. E. (1975). “Survey of the Sipuncula of the coral and beach rock communities of the Caribbean Sea,” in Proceedings of the International Symposium of Sipuncula and Echiura, eds M. E. Rice and M. Todorovic (Belgrade: Naucno Delo Press), 35–49.
Rice, M. E., and MacIntyre, I. G. (1982). Distribution of Sipuncula in the coral reef community, Carrie Bow Cay, Belize. Smiths. Contribut. Mar. Sci. 12, 311–320.
Rodriguez, E. A., Campbell, R. E., Lin, J. Y., Lin, M. Z., Miyawaki, A., Palmer, A. E., et al. (2017). The growing and glowing toolbox of fluorescent and photoactive proteins. Trends Biochem. Sci. 42, 111–129. doi: 10.1016/j.tibs.2016.09.010
Schultz, D. T., Kotlobay, A. A., Ziganshin, R., Bannikov, A., Markina, N. M., Chepurnyh, T. V., et al. (2018). Luciferase of the Japanese syllid polychaete Odontosyllis umdecimdonta. Biochem. Biophys. Res. Commun. 502, 318–323. doi: 10.1016/j.bbrc.2018.05.135
Schulze, A., Cutler, E. B., and Giribet, G. (2007). Phylogeny of sipunculan worms: a combined analysis of four gene regions and morphology. Mol. Phylogenet. Evol. 42, 171–192. doi: 10.1016/j.ympev.2006.06.012
Schulze, A., and Kawauchi, G. Y. (2021). How many Sipunculan species are hiding in our oceans? Diversity 13:43. doi: 10.3390/d13020043
Schulze, A., and Rice, M. E. (2004). Sipunculan diversity of Twin Cays, Belize, with a key to the species. Atoll Res. Bull. 521, 1–9.
Schulze, A., and Rice, M. E. (2009). Nephasoma pellucidum – a model species for sipunculan development? Smiths. Contribut. Mar. Sci. 38, 209–217.
Schulze, A., Boyle, M. J., and Kawauchi, G. Y. (2019). “Chapter 6.1: Sipuncula,” in Annelida, eds G. Purschke, M. Böggemann, and W. Westheide (Berlin: De Gruyter), 177–201.
Shimomura, O., and Johnson, F. H. (1968). Chaetopterus photoprotein: crystallization and cofactor requirements for bioluminescence. Science 159, 1239–1240. doi: 10.1126/science.159.3820.1239
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Struck, T., Schult, N., Kusen, T., Hickman, E., Bleidorn, C., McHugh, D., et al. (2007). Annelid phylogeny and the status of Sipuncula and Echiura. BMC Evolut. Biol. 7:57.
Tessler, M., Gaffney, J. P., Oliveira, A. G., Guarnaccia, A., Dobi, K. C., Gujarati, N. A., et al. (2020). A putative chordate luciferase from a cosmopolitan tunicate indicates convergent bioluminescence evolution across phyla. Sci. Rep. 10:17724. doi: 10.1038/s41598-020-73446-w
Vacher, M., Fdez Galván, I., Ding, B. W., Schramm, S., Berraud-Pache, R., Naumov, P., et al. (2018). Chemi- and bioluminescence of cyclic peroxides. Chem. Rev. 118, 6927–6974. doi: 10.1021/acs.chemrev.7b00649
Verdes, A., and Gruber, D. F. (2017). Glowing worms: biological, chemical, and functional diversity of bioluminescent annelids. Integr. Comparat. Biol. 57, 18–32. doi: 10.1093/icb/icx017
Waldenmaier, H. E., Oliveira, A. G., and Stevani, C. V. (2012). Thoughts on the diversity of convergent evolution of bioluminescence on earth. Int. J. Astrobiol. 11, 335–343. doi: 10.1017/S1473550412000146
Weigert, A., Helm, C., Meyer, M., Nickel, B., Arendt, D., Hausdorf, B., et al. (2014). Illuminating the base of the annelid tree using transcriptomics. Mol. Biol. Evol. 31, 1391–1401.
Widder, E. A. (2010). Bioluminescence in the ocean: origins of biological, chemical, and ecological diversity. Science 328:704. doi: 10.1126/science.1174269
Zhbannikov, I. Y., Hunter, S. S., Foster, J. A., and Settles, M. L. (2017). “SeqyClean: a pipeline for high-throughput sequence data preprocessing,” in Proceedings of the 8th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, (Boston, MA: Association for Computing Machinery).
Keywords: annelida, luciferin, luciferase, photoprotein, marine invertebrate
Citation: Oliveira AG, Amaral DT, Hannon MC and Schulze A (2021) First Record of Bioluminescence in a Sipunculan Worm. Front. Mar. Sci. 8:762706. doi: 10.3389/fmars.2021.762706
Received: 22 August 2021; Accepted: 28 September 2021;
Published: 20 October 2021.
Edited by:
Jerome Mallefet, Catholic University of Louvain, BelgiumReviewed by:
Jérôme Delroisse, University of Mons, BelgiumCarol Simon, Stellenbosch University, South Africa
Copyright © 2021 Oliveira, Amaral, Hannon and Schulze. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anderson G. Oliveira, anderson.garbuglio@usp.br; Anja Schulze, schulzea@tamug.edu
 Anderson G. Oliveira
Anderson G. Oliveira Danilo T. Amaral
Danilo T. Amaral Mary Colleen Hannon
Mary Colleen Hannon Anja Schulze
Anja Schulze