- 1Department of Plant Protection, College of Agriculture, Yangtze University, Jingzhou, China
- 2National Key Laboratory of Wheat and Maize Crop Science, Agronomy College, Henan Agricultural University, Zhengzhou, China
Plants of the Iris genus have been widely cultivated because of their medicinal, ornamental, and economic values. It commonly suffers from Alternaria leaf spot or blight disease leading to considerable losses for their commercial values. During an investigation of 14 provinces or municipalities of China from 2014 to 2022, a total of 122 Alternaria strains in section Alternaria were obtained from diseased leaves of Iris spp.. Among them, 12 representative strains were selected and identified based on morphological characterization and multi-locus phylogenetic analysis, which encompassed the internal transcribed spacer of rDNA region (ITS), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), translation elongation factor 1 alpha (TEF1), RNA polymerase second largest subunit (RPB2), Alternaria major allergen gene (Alt a 1), an anonymous gene region (OPA10-2), and endopolygalacturonase gene (EndoPG). The strains comprised two known species of A. alternata and A. iridicola, and two new species of A. setosae and A. tectorum, which were described and illustrated here. Their pathogenicity evaluated on Iris setosa indicated that all the strains could induce typical Alternaria leaf spot or blight symptoms. The results showed that the virulence was variable among those four species, from which A. tectorum sp. nov. was the most virulent one, followed by A. setosae sp. nov., A. iridicola and A. alternata.
Introduction
The Iris Linn. is the largest genus in the Iridaceae containing about 300 species in worldwide (Xiang et al., 2020). It is one of the most extensively cultivated plants in modern landscaping because it is a great ground cover material for urban greening (Zhao et al., 2000; Clair, 2005; Li et al., 2020). In China, the plants have been widely grown as a year-round ornamental with various large and colorful flowers (Yang et al., 2017). Some Iris species are rich in beneficial chemicals, served as effective pharmaceutical ingredient to treat various diseases including cancer, inflammation, bacterial and viral infections (Tahara et al., 1991; Huang, 2004; Meng et al., 2016). In addition, Iris plants are also utilized in the perfume and cosmetic industries considering the fragrance reasonability (Wang et al., 2010).
During the cultivation of Iris spp., the plants suffer from various diseases induced by fungal, bacterial, viral, and nematode pathogens (Umesha et al., 2010; Lu et al., 2016; Wang et al., 2020; Wang et al., 2022). Pathogenic fungal species have been commonly found in connection with the genera of Alternaria, Botrytis, Colletotrichum, Fusarium, Harzia and Puccinia (Bobev, 2009; Liu et al., 2016; Schultes et al., 2017; Choi et al., 2019; Wang et al., 2022). Leaf spot and blight diseases caused by Alternaria species in sect. Alternaria is prevalent on Iris plants worldwide presenting the typical symptoms of brown spot with a yellow halo or blighted leaf, which greatly reduce their ornamental values. Alternaria iridicola in China, Japan, and USA (Zhang, 2003; Simmons, 2007; Nishikawa and Nakashima, 2020), A. iridiaustralis in Australia, China, and New Zealand (Simmons, 2007; Luo et al., 2018), A. tenuissima in China (Zhang, 2003; Sun et al., 2019; Li et al., 2020) have been reported as fungal pathogens on Iris plants.
The Alternaria is a ubiquitous fungus comprising many destructive plant pathogens, which can result in economic losses on a large number of significant agronomic crops and ornamentals (Thomma, 2003; Kahl et al., 2015). There has been a longtime controversy on the classification criteria of Alternaria (Lawrence et al., 2016). Simmons proposed a reasonable standard for the morphological taxonomy of Alternaria species based on sporulation patterns and conidial morphology, by which around 280 species were described and summarized into two sections of large-spored and small-spored of Alternaria (Simmons, 2007). Since the 20th century, molecular taxonomic structure has been developed and applied to identify Alternaria species with various gene fragments (Pryor and Gilbertson, 2000; Hong et al., 2005; Lawrence et al., 2012, 2013; Woudenberg et al., 2013, 2014; He et al., 2021). Recently, the multi-locus phylogenetic analysis plays an important role in assisting Alternaria classification (Woudenberg et al., 2015; Zheng et al., 2015; Sofie et al., 2017; Cheng et al., 2022). It separates Alternaria into 29 sections and 7 monotypic lineages (Woudenberg et al., 2013; Marin-Felix et al., 2019; Bessadat et al., 2020; Gannibal et al., 2022; Huang et al., 2022). Besides, in combination with morphology and molecular approaches is commonly used to determine the Alternaria up to species levels (Lawrence et al., 2016; Gannibal, 2019; Bessadat et al., 2021; Zhao et al., 2022).
One of the large Alternaria sections, sect. Alternaria contains most of the small-spored Alternaria species with concatenated conidia including about 60 morphological or host-specific species (Simmons, 2007; Woudenberg et al., 2015), which are frequently encountered on the disease leaf samples of Iris plants. To know the species population associated with Iris plants in China, a large-scale sample collection and fungal isolation had been conducted from 2014 to 2022, from which a total of 122 strains of sect. Alternaria were obtained. To comprehend their species levels, this study aims to identify the species using morphological traits and multi-locus phylogenetic analysis. In addition, their variety of virulence is assessed on Iris setosa.
Materials and methods
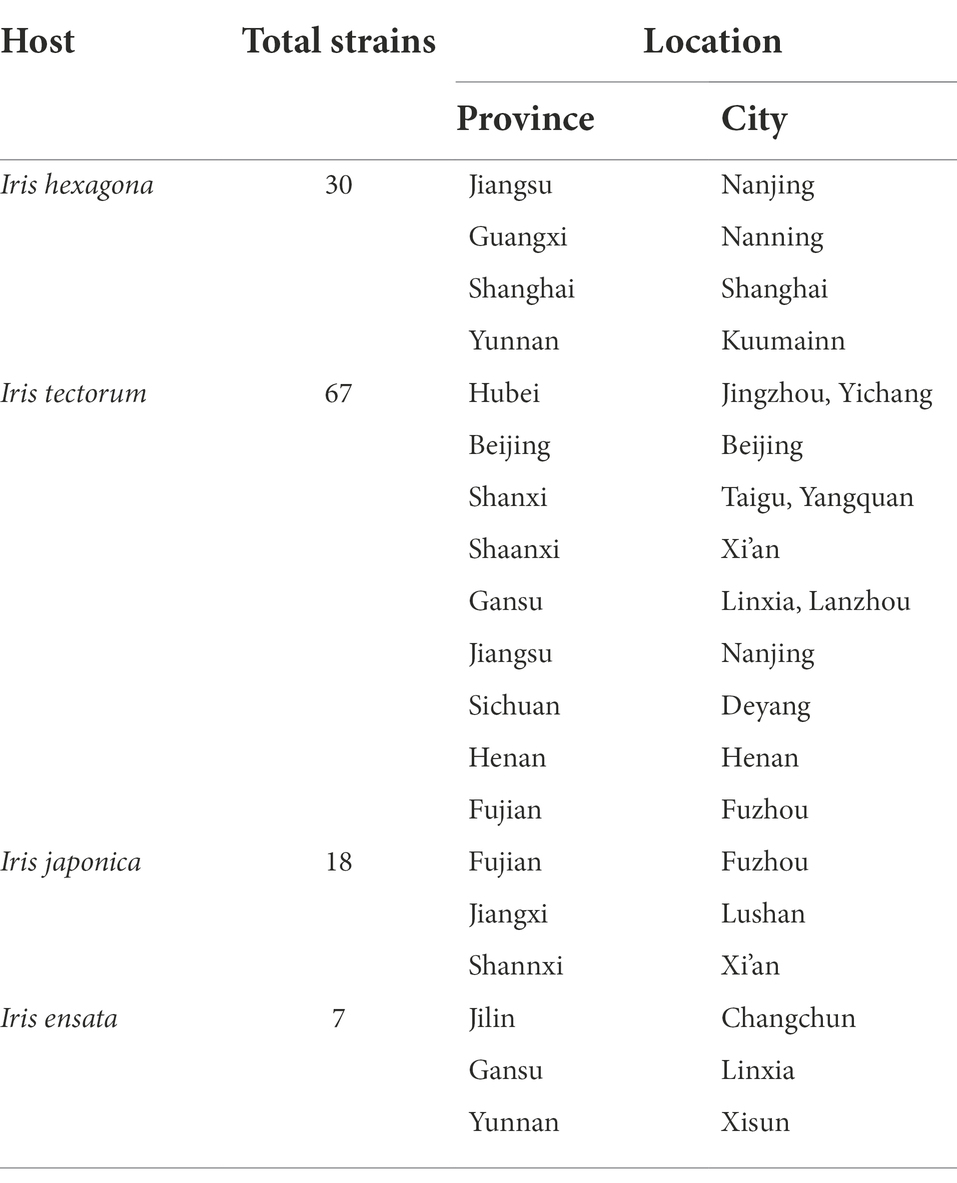
Sampling and isolation
During a large-scale investigation on Alternaria species in sect. Alternaria associated with Iris spp. in China (Figure 1), diseased leaf samples were collected from 14 provinces or municipalities (Table 1). For the isolation of Alternaria, the samples were cut into small pieces, placed on moist filter papers in Petri dishes and incubated at 25°C for the sporulation (Liu et al., 2019). The samples were observed using a stereomicroscope. The single spore was picked by a sterile glass needle and inoculated onto potato dextrose agar (PDA: Difco, Montreal, Canada). A total of 122 strains were obtained and kept into test-tube slants stored at 4°C in the Fungal Herbarium of Yangtze University (YZU), Jingzhou, Hubei, China. Twelve representative strains (Table 2) representing different species were selected for the present study after pre-morphological, pre-phylogenetic, and pre-pathogenicity assays.
Morphology
The Alternaria strains were cultured on PDA at 25°C for 7 days in darkness to determine the cultural features. To examine the conidial morphology, fresh mycelia were grown on potato carrot agar (PCA) and V8 juice agar (V8A) media, then incubated at 22°C with a light period of 8 h light/16 h dark (Simmons, 2007). After 7 days, morphological characteristics of sporulation patterns, conidiophores and conidia were visualized and photographed with a Nikon Eclipse Ni-U microscope system (Nikon, Japan). Fifty randomly selected conidia for each strain were observed and measured to determine the morphology.
DNA extraction and PCR amplification
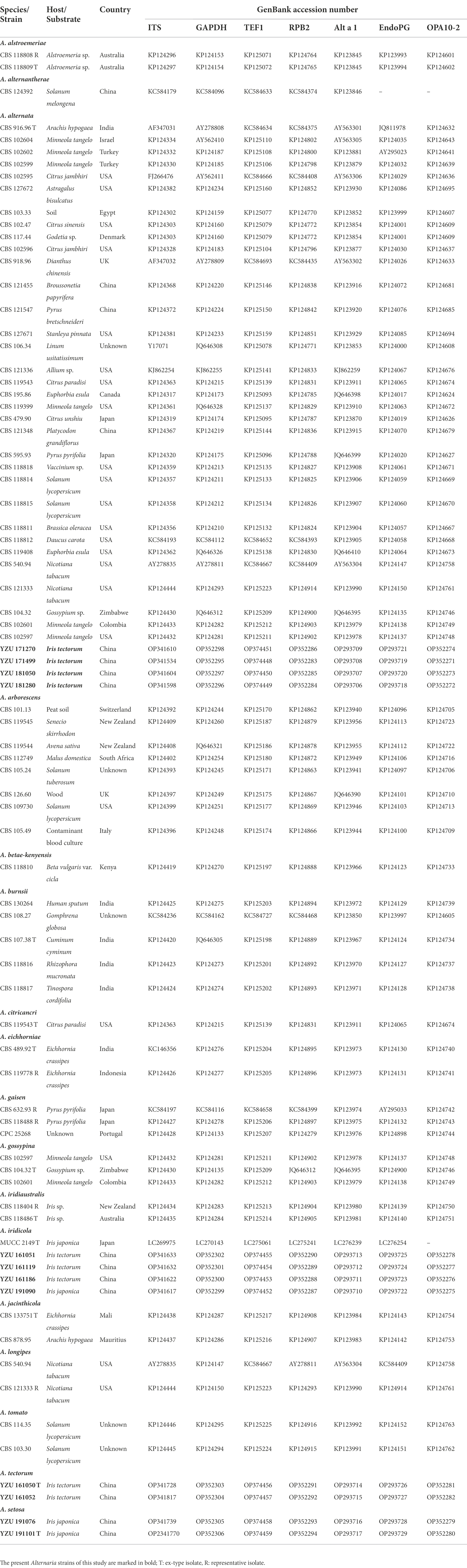
Genomic DNA was extracted from mycelium scraped from the surface of 5-day-old colonies on PDA using the CTAB method described in Watanabe et al. (2010). Seven gene regions including internal transcribed spacer of rDNA region (ITS), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), translation elongation factor 1 alpha (TEF1), RNA polymerase second largest subunit (RPB2), Alternaria major allergen gene (Alt a 1), an anonymous gene region (OPA10-2), and endopolygalacturonase (EndoPG) were used for phylogenic analysis. The PCR amplifications were performed with primer pairs of ITS5/ITS4 (White et al., 1990), gpd1/gpd2 (Berbee et al., 1999), EF1-728F/EF1-986R (Carbone and Kohn, 1999), RPB2-5F/RPB2-7cR (Liu et al., 1999), Alt-for/Alt-rev (Hong et al., 2005), OPA10-2 l/OPA10-2R (Andrew et al., 2009) and PG3/PG2b (Andrew et al., 2009), respectively. A 25-μL PCR mixture contained 21 μl 1.1× Taq PCR Star Mix (TSINGKE, Beijing, China), 2 μl template DNA, and 1 μl of each primer was performed in a BIO-RAD T100 thermo cycler. The amplified program for PCR amplifications of the seven gene regions was referenced from Woudenberg et al. (2015). Successful amplified products were purified and sequenced by TSINGKE company (Beijing, China). The obtained sequences for each gene were deposited in GenBank1 with the accession numbers indicated in Table 2.
Phylogenetic analysis
The ITS, GAPDH, TEF1, RPB2, Alt a 1, OPA10-2 and EndoPG gene sequences were launched for BLAST searches in NCBI.2 Their relevant sequences were retrieved from GenBank database and referenced from Woudenberg et al. (2015; Table 2). Alternaria alternantherae (CBS 124392) from Alternaria section of Alternantherae was used as outgroup taxon (Table 2). Sequences were aligned and edited by the program of PHYDIT v3.2 (Chun, 1995). The seven gene sequences were concatenated and edited manually in MEGA v.7.0.26 (Kumar et al., 2016). Phylogenetic trees were constructed based on Bayesian inference (BI) analysis using MrBayes v.3.1.2 (Ronquist and Huelsenbeck, 2003), and maximum likelihood (ML) method using RAxML v.7.2.8 (Stamatakis, 2006). The best-fit model for the data was calculated by the Akaike Information Criterion (AIC) using MrModelTest v. 2.3 (Posada and Crandall, 1998). The Bayesian analysis (MrBayes v. 3.2.1; Ronquist et al., 2012) of two simultaneous Markov Chain Monte Carlo (MCMC) chains were run from random trees for 10, 000, 000 generations and sampled every 100 th generations. The first 25% of the samples were discarded as the burn-in and the run was automatically stopped when the average standard deviation of split frequencies reached below 0.01. The phylogram was plotted and edited in Figtree v.1.3.1 (Rambaut and Drummond, 2010). The branch support for the ML analysis was assessed with 1,000 replicates.
Pathogenicity tests
Iris setosa was commonly cultivated as an ornamental or medicinal plant in China. The plants were purchased from the local market and transplanted into clean pots with organic sterile soil grown in greenhouse under a light period (12 h) at 25°C for 2 months. The test strains were cultured on PDA at 25°C for 5 days, and a diameter of 6 mm disc was removed from the edge of colonies. It was inoculated on wounded living leaves of I. setosa helped with a fine sterile needle (eight punctures). Clean PDA discs were used as controls. To ensure an accuracy of pathogenicity assessment, a consistent treatment method was adopted for each assay. For each strain, two plants were inoculated with four sites, which were replicated for three times. To confirm the Koch’s law, the pathogen was re-isolated from the diseased symptoms and compared with the original strains based on morphology. The disease incidence was recorded, and the lesion size (LS = the maximum lesion length) was measured after 7 days. The LS values were the mean value of three replicates ± standard deviation. The least significant difference test (p < 0.05) was analyzed by IBM SPSS Statistics 23.
Results
Phylogenetic analysis
On the basis of BLAST searches, all 122 Alternaria strains were belonged to sect. Alternaria. Phylogenetic analyses using the combined dataset of the seven gene sequences (ITS, GAPDH, TEF1, RPB2, Alt a 1, OPA10-2 and EndoPG) were conducted including 79 strains of sect. Alternaria (Table 2), which comprised 3,340 characters (470 from ITS,534 from GAPDH,240 from TEF1,603 from RPB2,429 from Alt a 1,622 from OPA10-2,442 from EndoPG). The resulted topologies of BI and ML analysis were similar to each other, and the ML tree was shown in Figure 2 for basal phylogeny. It showed that strains of YZU 171270, YZU 171499, YZU 181050 and YZU 181280 fell into four different subclades of A. alternata. Strains of YZU 161051, YZU 161186, YZU 161119 and YZU 191090 were grouped well together with A. iridicola supported by values of 1.0/100% (PP/BS). Two strains of YZU 191101 and YZU 191076 fell into a distinct single lineage (PP/BS = 1.0/99%) close to A. gaisen supported by the values of 1.0/100% (PP/BS), which could be considered as a new species. The remaining two strains of YZU 161050 and YZU 161052 also formed an individual branch with high PP/BS values of 1.0/100% representing a new species in the phylogram, sister to A. iridicola (PP/BS = 0.99/98%; Figure 2). Besides, the four strains, A. gaisen and A. iridicola were grouped together supported with medium PP/BS values of 0.8/65%.
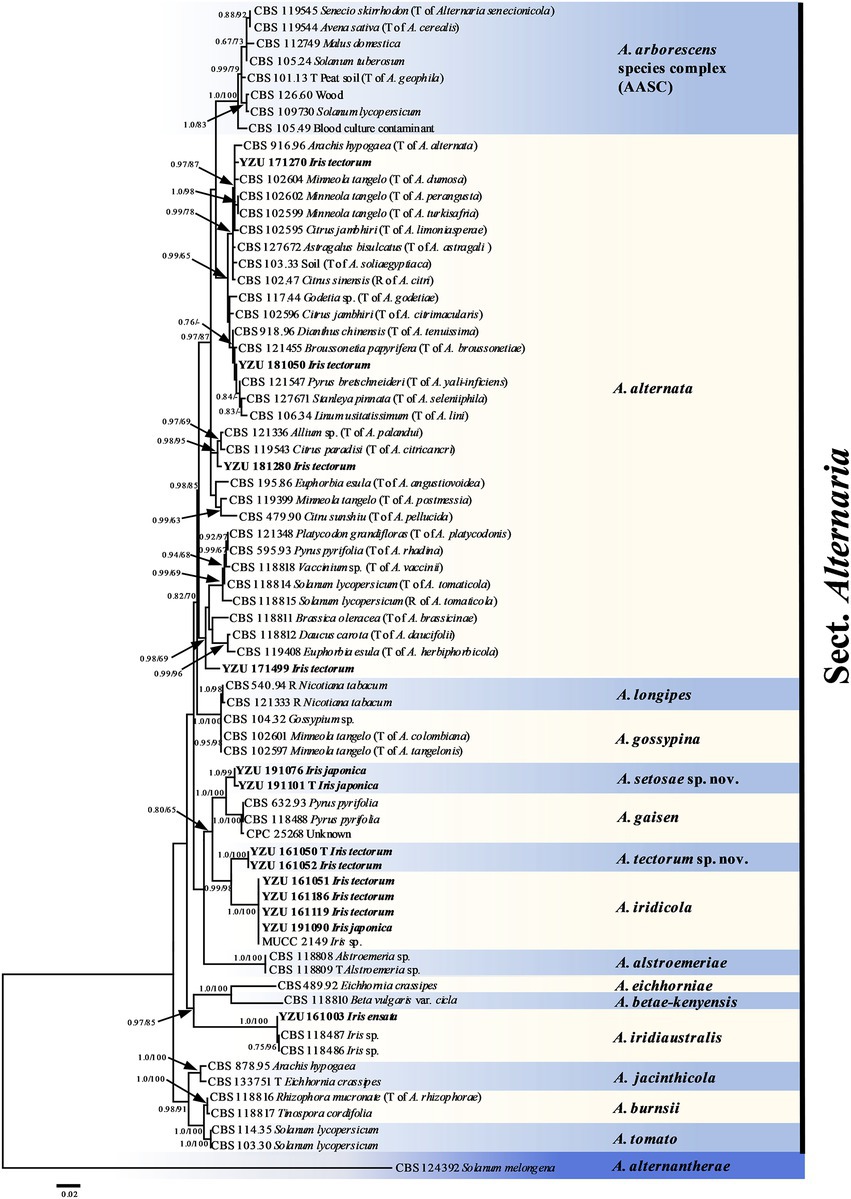
Figure 2. Phylogenetic tree based on the combined gene sequences of ITS, GAPDH, TEF1, RPB2, Alt a 1, OPA10-2 L and EndoPG generated from Alternaria spp. on Iris plants. The Bayesian posterior probabilities (PP > 0.60) and maximum likelihood bootstrap values (BS > 60%) are given at the nodes (PP/BS). Examined present strains are in bold.
Morphology
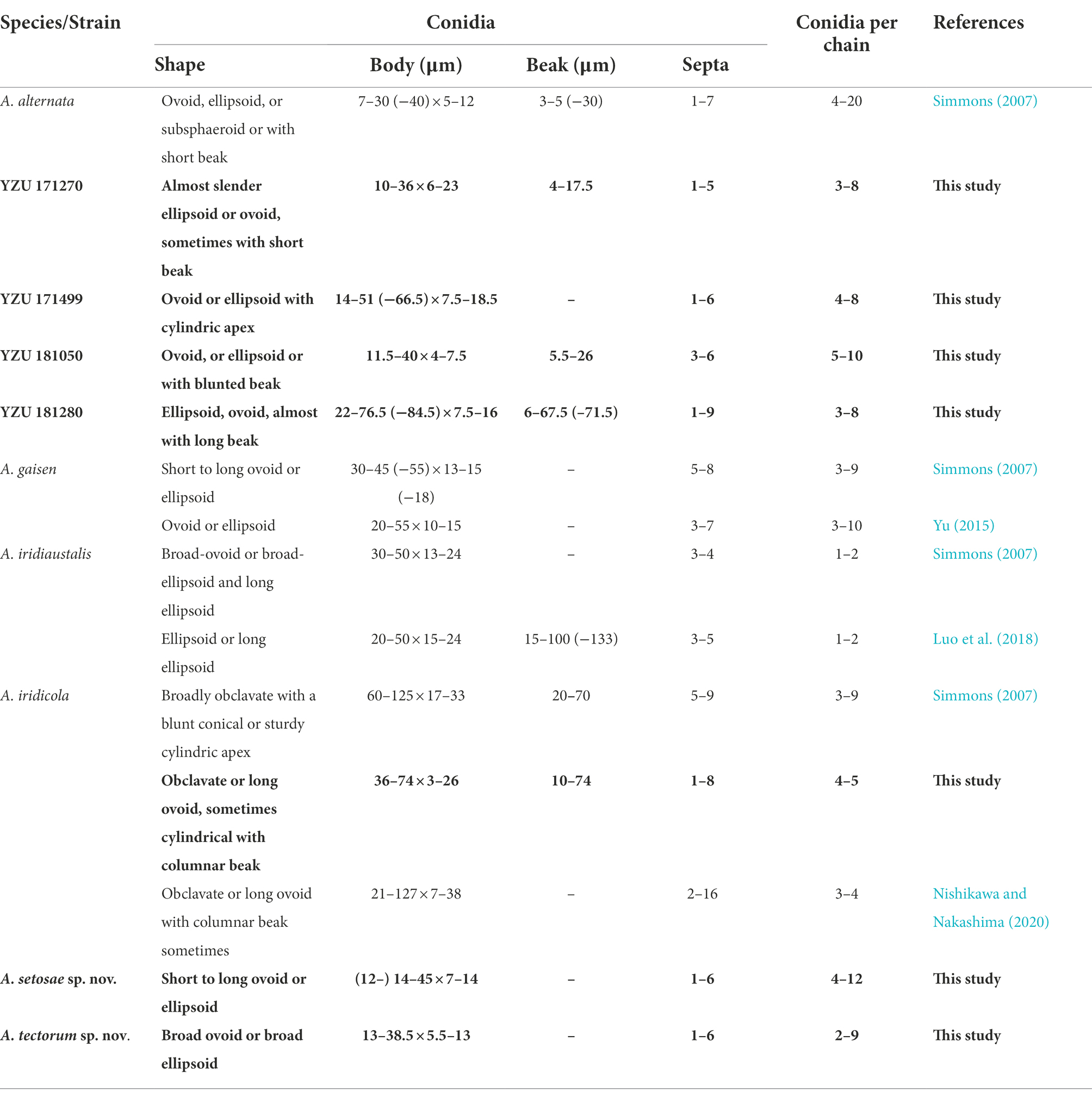
According to morphological traits, the present Alternaria strains associated with Iris spp. were A. alternata (Figure 3) and A. iridicola (Figure 4) and two new species of sect. Alternaria (Figures 5, 6; Table 3). The two new species were illustrated and described in this study.
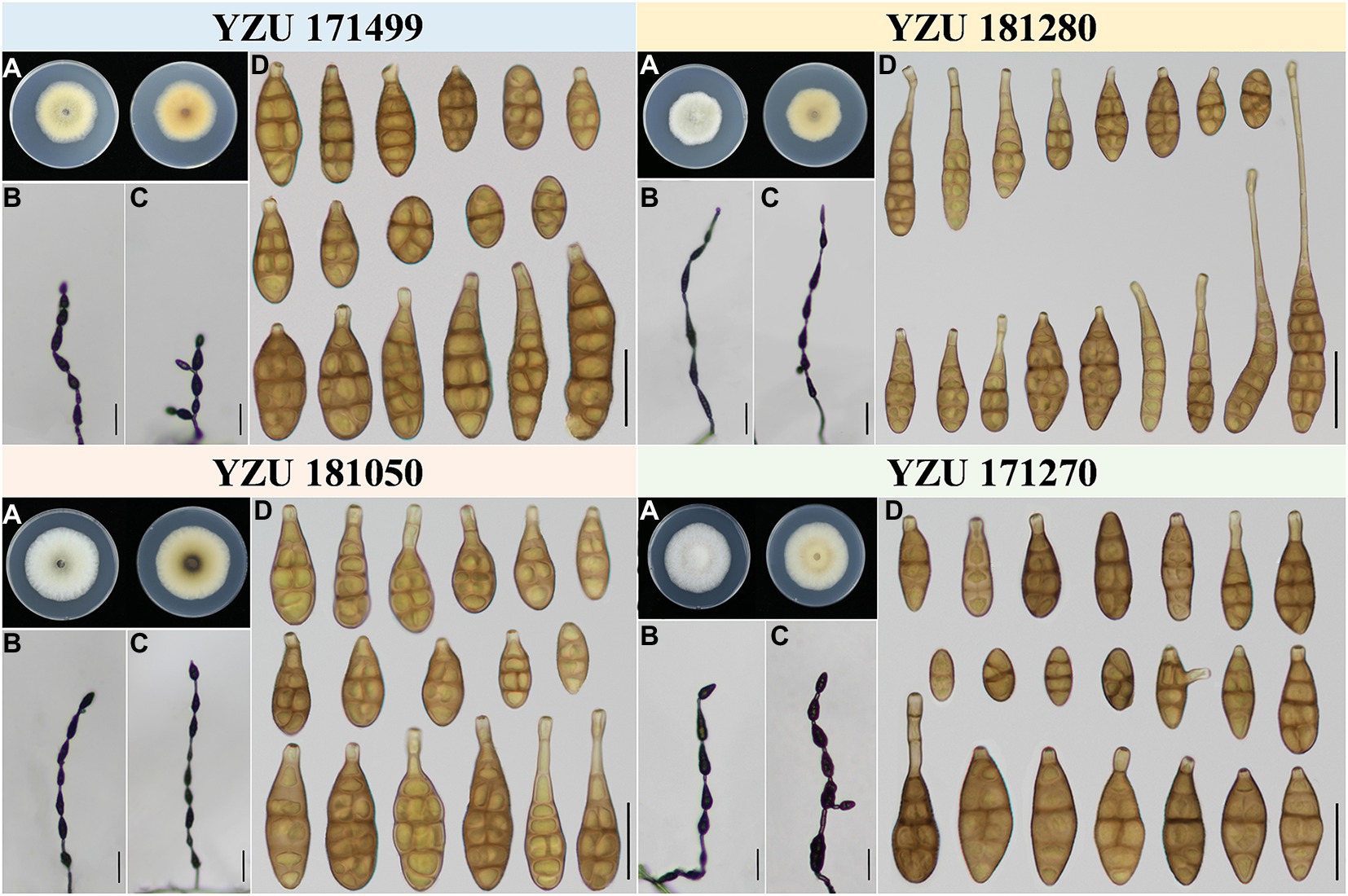
Figure 3. Morphology of the represent four strains of A. alternata. (A) Colony phenotype (on PDA for 7 days at 25°C); (B,C) Sporulation patterns (on PCA at 22°C); (D) Conidia Bars: B,C = 50 μm; D = 25 μm.
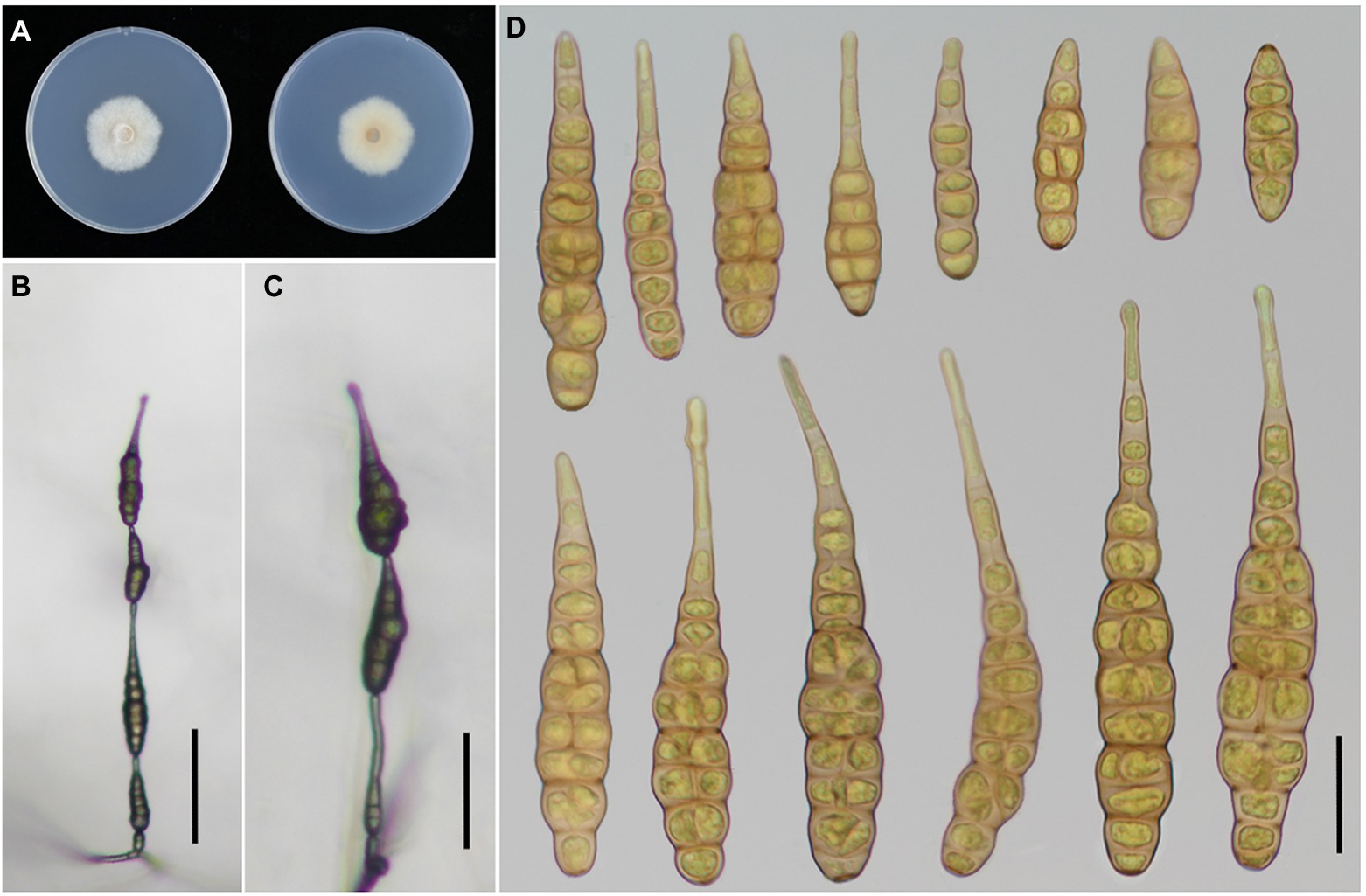
Figure 4. Morphology of Alternaria iridicola. (A) Colony phenotype (on PDA for 7 days at 25°C); (B,C) Sporulation patterns (on PCA at 22°C); (D) Conidia Bars: B,C = 50 μm; D = 25 μm.
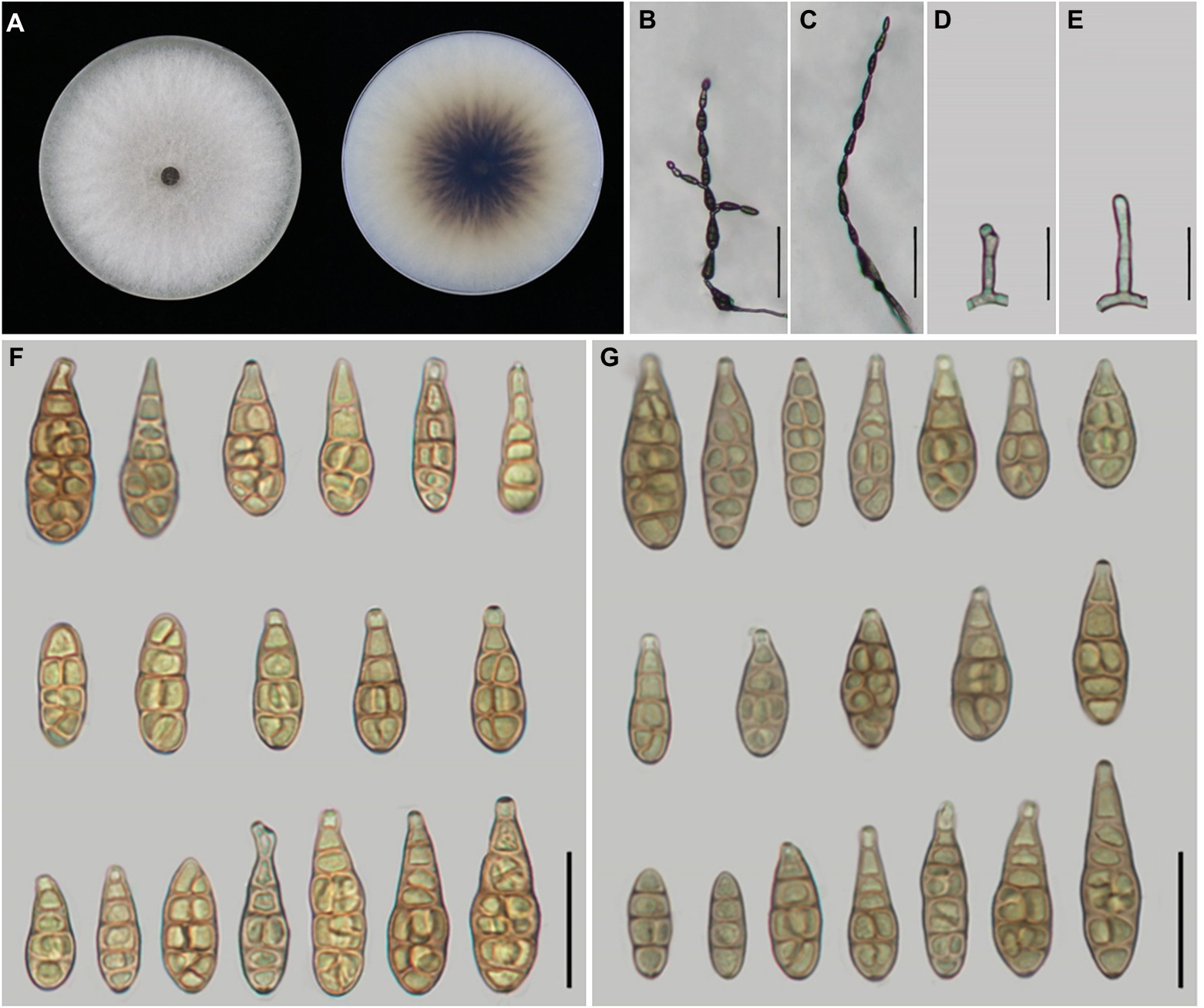
Figure 5. Morphology of Alternaria setosae sp. nov. (A) Colony phenotype (on PDA for 7 days at 25°C); (B,C) Sporulation patterns (on PCA at 22°C); (D,E) Conidiophores; (F) Conidia (on PCA at 22°C); (G) Conidia (on V8A at 22°C). Bars: B,C = 50 μm; D–G = 25 μm.
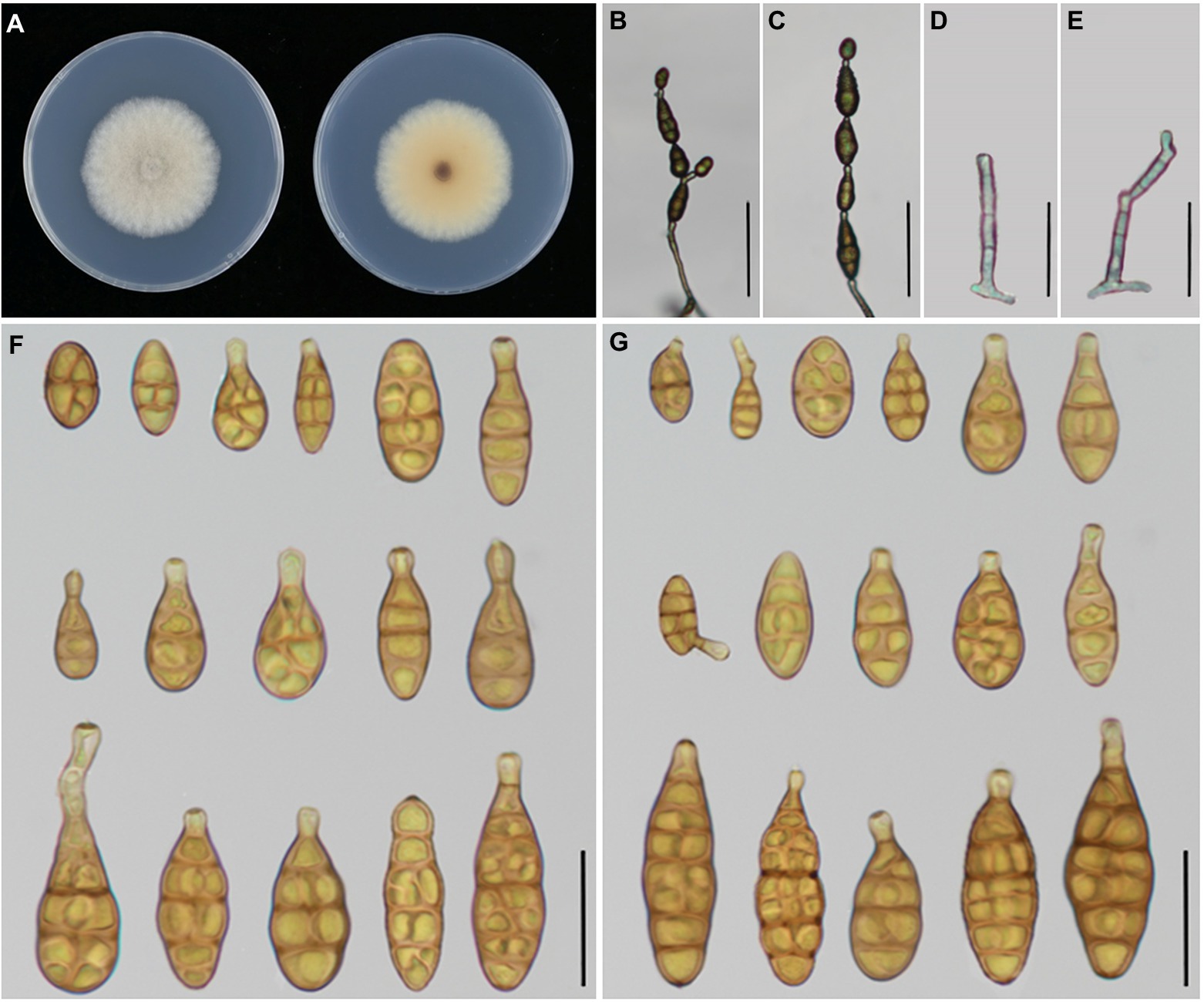
Figure 6. Morphology of Alternaria tectorum sp. nov. (A) Colony phenotype (on PDA for 7 days at 25°C); (B,C) Sporulation patterns (on PCA at 22°C); (D,E) Conidiophores; (F) Conidia (on PCA at 22°C); (G) Conidia (on V8A at 22°C). Bars: B,C = 50 μm; D–G = 25 μm.
Taxonomy
Alternaria setosae Y.N. GOU & J.X. Deng, sp. nov. Figure 5
MycoBank No.: 845326
Etymology: In reference to the pathogenic host species name, Iris setosa.
Typifification: China, Fujian Province, Fuzhou City, from leaf spot of Iris japonica, 15 April 2019, J.X. Deng, (YZU-H−0046, holotype), ex-type culture YZU 191101.
Description: Colonies on PDA circular, light cottony and white to off-white in the center, villiform with white at the edge, reverse dark brown at centers, with scattering shape, 83.4–84.8 mm in diam., at 25°C for 7 days. On PCA, conidiophores arising from substrate or lateral of aerial hyphae, straight or curved, smooth-walled, septate, pale brown; (12.5–)15–40 × 3–5 μm (av.: 24.5 × 4 μm); conidia 4–12 units per chain, medium yellow-brown with almost smooth-walled, short to long-ovoid or ellipsoid, (12–)14–45 × 7–14 μm (av.: 31.5 × 10.5 μm),1–6 transverse septa,0–2(−3) longitudinal septa. On V8A, conidiophores straight or curved, smooth-walled, septate, 18–43(−50.5) × 3.5–5 μm (av.: 31 × 4 μm), conidia 4–11 units in a chain, medium yellow-brown, almost smooth to conspicuously elliptoid overall, 20–43 × 7–12.5 μm (av.: 30 × 9.5 μm), 2–7 transverse septa, 0–2(−3) longitudinal septa.
Notes: Phylogenetic analysis reveals that the species is sister to A. gaisen on the basis of a combined dataset of ITS, GAPDH, TEF1, RPB2, Alt a 1, OPA10-2 and EndoPG gene regions. After a nucleotide pairwise comparison with A. gaisen in those seven regions, there are 1/470 bp, 1/534 bp, 3/240 bp, 3/603 bp, 0/429 bp, 16/622 bp and 0/442 bp site differences, respectively. Morphologically, this species can be differentiated by producing smaller conidia with less transverse septa (Table 3). It also readily distinct to A. tectorum sp. nov. and A. iridicola in sporulation pattern and conidial size (Table 3).
Alternaria tectorum Y.N. GOU & J.X. Deng, sp. nov. Figure 6
MycoBank No.: 845325
Etymology: In reference to the host species name, Iris tectorum.
Typifification: China, Hubei Province, Jingzhou City, from leaf spot of Iris tectorum, 21 May 2016, J.X. Deng, (YZU-H−0025, holotype), ex-type culture YZU 161050.
Description: Colonies on PDA circular, cottony with dense hyphae, off-white, reverse buff in the center, with white margin, 50–52 mm in diam., at 25°C for 7 days. On PCA, conidiophores arising from substrate, straight to slightly curved, septate, pale to dark brown, 32–58.5(−62.5) × 3.5–4.8 μm (av.: 45.5 × 4 μm); conidia 1–6 in a chain, solitary or straight in chains with few branches, broad-ovoid or broad-ellipsoid, smooth-walled, 13–38.5 × 5.5–13 μm (av.: 23.5 × 9 μm), 0–6 transverse septa, 0–2(−3) longitudinal septa, On V8A, conidiophores straight to slightly curved, septate, pale to dark brown, 31.5–54.5 × 3.5–4.75 μm (av.: 41.5 × 4 μm),conidia 2–7 per chain, pale to medium yellowish brown, smooth walled, broad-ovoid or broad-ellipsoid, 13.5–37.5 × 5.5–13.5 (av.: 26 × 9.5 μm), 1–7 transverse septa, 0–2(−3) longitudinal septa.
Notes: Phylogenetic analysis shows that the species (13 –38.5 × 5.5 –13 μm in body size) falls in an individual clade with high PP/BS values of 1.0/100% representing a new species in the phylogram, sister to A. iridicola (47 -87 × 15 -27 μm in size), but it can be significantly distinguished by the conidial size and shape (Table 3). Comparing with A. iridicola, the present species contains 1/442 bp, 16/603 bp and 37/622 bp nucleotide differences in EndoPG, RPB2 and OPA10-2 gene sequences, respectively.
Pathogenicity tests
Pathogenicity tests inoculated with those 12 Alternaria strains for 7 days, the results showed that all strains were 100% pathogenic to Iris setosa (Table 4; Figure 7). When inoculated with mycelium discs, the small spot was normally appeared after 4 days on the living leaves. Then it expanded into nearly round to long elliptic, dark brown necrotic lesions coupling with a narrow yellow halo around the periphery of the spot. Besides, the necrotic part of spot is easy to rupture and form perforation. For some strains (A. tectorum), the spots spread quickly until most of the leaves becoming withered causing blight. The symptoms were identical to those in the fields (Figure 1). The virulence among the species was variable (Table 4). Strains of A. tectorum sp. nov. showed the most virulent to I. setosa with the lesion size (LS) around 48 to 49 mm, followed by A. setosae sp. nov. (LS around 26–27), then A. iridicola and A. alternata. There were no symptoms on the control leaves. To verify Koch’s rules, the re-isolation of the causal pathogen was performed, which was consistent with the inoculated strains.
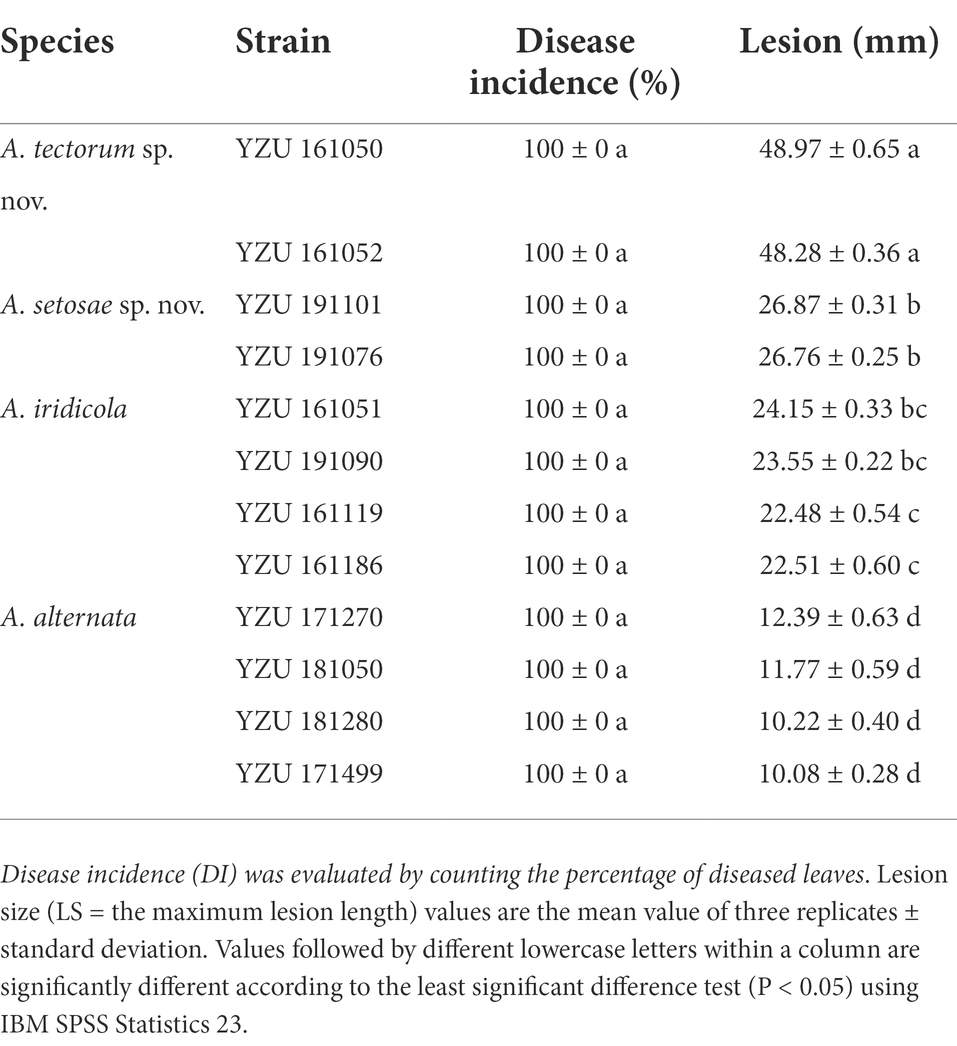
Table 4. Disease incidence and lesion size on Iris setosa induced by the present Alternaria species.
Discussion
The sect. Alternaria is found containing 17 species and a species complex of A. arborescens (AASC; Woudenberg et al., 2015; Li et al., 2022). After a large-scale sample collection of Iris plants in China, two known species (A. alternata and A. iridicola) and two novel species in sect. Alternaria were detected in this study. In addition, the two new species also identified and described as A. setosae sp. nov. and A. tectorum sp. nov. based on the morphological characteristics and multi-locus phylogenetic analyses.
Alternaria alternata is one kind of small-spored Alternaria, which taxonomy is controversial due to the similar conidial morphology (Zhang, 2003; Simmons, 2007). Later, it is classified as one species based on multi-gene phylogenetic analyses, encompassing 35 morphospecies identified by Simmons (2007) which could not reliably distinguish those species (Woudenberg et al., 2015). Meanwhile, it resulted in confusion to the related taxonomic works on their classification. Morphologically, strain YZU 171270 grouped in a sub-clade containing the type strain of A. alternata (Figure 2) can form long catenulate chain with branching 1 to 3, while strain YZU 181050 in its sister sub-clade containing the type strain of A. tenuissima can sporulate with long straight conidial chains without branch. The results agreed with Simmons (2007) for the descriptions of A. alternata and A. tenuissima. In China, A. tenuissima has been reported on I. tectorum in Qingdao city (Sun et al., 2019) and I. ensata in Anqing city (Li et al., 2020), whose sporulation patterns are not provided. Although sporulation phenotype cannot accurately reflect the evolutionary relationship, it still is of great value for subgroup classification of A. alternata. Besides, the present strains falling into different clade or subclade (Figure 2) are highly variable (Figure 3). For example, both strains YZU 181050 and YZU 181280 are similar in sporulation, but strain YZU 181280 is characterized by comprising larger conidia with blunted beak up to 71.5 μm long (Figure 3). The morphological boundary of A. alternata has not been clearly defined yet and how to classify them accurately needs further study (Aung et al., 2020).
Alternaria iridicola has been recognized as a large-spored Alternaria (ca. 60–125 × 17–33 μm in size) comprising broadly obclavate conidia with a blunt conical or sturdy cylindric apex, which is a host specific species to Iris plants (Simmons, 2007). Gannibal and Lawrence (2018) considered that A. iridicola shares traits with both sect. Panax and Porri of Alternaria, which was not easily distinguished in terms of section affiliation. Latterly, this species is the only large-spored Alternaria in small-spored sect. Alternaria according to multi-locus phylogenetic analysis (Nishikawa and Nakashima, 2020). Nishikawa and Nakashima (2020) took experiments to assess the host range of A. iridicola, for which I. laevigata and I. hollandica were proved host related, except I. ensata. The species was firstly reported causing leaf spot disease on I. tectorum from Xinxiang city of China in 2015 (Zhai et al., 2015). In this study, I. japonica is a new host for A. iridicola in China, which also can infect I. setosa.
In this study, two new species of A. setosae and A. tectorum are grouped with A. gaisen and A. iridicola in a clade, sister to A. alstroemeriae. The morphological comparisons are listed in Table 3. Compared with A. gaisen (30–45 (−55) × 13–15 (−18) μm with 5–8 septa), A. setosae sp. nov. is different by producing smaller conidia (14–45 × 7–14 μm) and less transverse septa (1–6; Simmons, 2007). In comparison with A. iridicola, A. tectorum sp. nov. is an obviously a small-spored species (Simmons, 2007). The results increase two members for sect. Alternaria. In the previous reports, A. iridiaustralis has been reported on Iris spp. in Australia and New Zealand (Simmons, 2007) and on Iris ensata as causal foliar pathogen in China (Luo et al., 2018), which is characterized by broad-ovoid or broad-ellipsoid and long ellipsoid conidia, obviously not like the present four species.
During the isolation of Alternaria from Iris plants, A. alternata like species are frequently observed on moisten cultured leaf tissues, such as A. alternata, A. iridicola, A. setosae sp. nov. and A. tectorum sp. nov.. Consequently, their virulence is assessed on wounded leaves of I. setosa, and considerable variation is observed among the species in the pathogenicity tests, from which A. tectorum sp. nov. shows the most pathogenic, followed by A. setosae sp. nov., A. iridicola, and finally A. alternata. The pathogenic mechanism comprises mechanical penetration, secreting degradation enzymes, metabolites, and toxins, which causes plant diseases (Meng et al., 2009). Iris plants are near to the ground growing, which suffers vulnerable damage caused by raining splashing, animals, and so on. When there are small wounds on Iris leaves, it is easier to attack by pathogens, which could significantly lower their economic and ornamental values. On the other hand, mycelia discs of the four species were inoculated on unwounded living leaves of Iris setosa for 14 days (data not shown). Alternaria tectorum sp. nov. (LS around 20 mm) and A. iridicola (LS 6 to 18 mm) can penetrate the leaves and induce symptoms. While A. alternata and A. setosae sp. nov. fail to penetrate and cause infection. The present results will provide experimental evidence and reference for the prevention and control of Iris leaf spot caused by Alternaria alternata like species.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
JD, SA, and YG did the study design. JD, SA, CH, and AH collected the diseased leaf samples. YG and SA performed the experiments and analyzed the data. YG wrote the manuscript. JD and YG contributed to manuscript revision, read, and approved the submitted version.
Funding
This study was financed by the National Natural Science Foundation of China (31400014 and 32270022).
Acknowledgments
The authors sincerely thank the editors and reviewers for taking their valuable time to review this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Andrew, M., Peever, T. L., and Pryor, B. M. (2009). An expanded multilocus phylogeny does not resolve morphological species within the small-spored Alternaria species complex. Mycologia 101, 95–109. doi: 10.2307/20619150
Aung, S., Liu, H. F., Pei, D. F., Lu, B. B., Oo, M. M., and Deng, J. X. (2020). Morphology and molecular characterization of a fungus from the Alternaria alternata species complex causing black spots on Pyrus sinkiangensis (koerle pear). Mycobiology 48, 233–239. doi: 10.1080/12298093.2020.1745476
Berbee, M. L., Pirseyedi, M., and Hubbard, S. (1999). Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia 91, 964–977. doi: 10.2307/3761627
Bessadat, N., Hamon, B., Bataillesimoneau, N., Mabrouk, K., and Simoneau, P. (2020). Alternaria telliensis sp. nov., a new species isolated from Solanaceae in Algeria. Phytotaxa 440, 89–100. doi: 10.11646/phytotaxa.440.2.1
Bessadat, N., Hamon, B., Bataillé-Simoneau, N., Mabrouk, K., and Simoneau, P. (2021). Characterization of new small-spored Alternaria species isolated from Solanaceae in Algeria. Life 11:1291. doi: 10.3390/life11121291
Bobev, S. (2009). Reference guide for the diseases of cultivated plants Makros Publication, Russia, 466.
Carbone, I., and Kohn, L. M. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91, 553–556. doi: 10.2307/3761358
Cheng, H., Zhao, L., Wei, X., Liu, X., Gao, G. Y., Deng, J. X., et al. (2022). Alternaria species causing leaf spot on hemp (Cannabis sativa) in northern China. Eur. J. Plant Pathol. 162, 957–970. doi: 10.1007/s10658-021-02450
Choi, I. Y., Choi, Y. J., Kim, J. Y., and Shin, H. D.. (2019). Identification of Puccinia iridis on Iris domestica in Korea. Korean J. Mycol. 47, 89–94. doi: 10.4489/KJM.20190011
Chun, J. (1995). Computer-assisted classification and identification of actinomycetes. Dissertation, Unversity of Newcastle.
Gannibal, P. D. (2019). New species and new findings in Russia of Alternaria sect. Gypsophilae. Mikologiya I Fitopatologiya 53, 10–16. doi: 10.1134/S0026364819010069
Gannibal, P. D., and Lawrence, D. P. (2018). Distribution of Alternaria species among sections. 5. Species producing conidia with many longitudinal septa. Mycotaxon 133, 285–291. doi: 10.5248/133.28
Gannibal, P. B., Orina, A. S., and Gasich, E. L. (2022). A new section for Alternaria helianthiinficiens found on sunflower and new asteraceous hosts in Russia. Mycol. Prog. 21. doi: 10.1007/s11557-022-01780-6
He, L., Cheng, H., Htun, A. A., Ge, H., Xia, Z. Z., Guo, J. W., et al. (2021). Phylogeny and taxonomy of two new Alternaria (Ascomycota: Pleosporaceae) species in section Gypsophilae from China. Mycol. Prog. 20, 355–363. doi: 10.1007/s11557-021-01676-x
Hong, S. G., Cramer, R. A., Lawrence, C. B., and Pryor, B. M. (2005). Alt a 1 allergen homologs from Alternaria and related taxa: analysis of phylogenetic content and secondary structure. Fungal Genet. Biol. 42, 119–129. doi: 10.1016/j.fgb.2004.10.009
Huang, S. Z. (2004). Evaluation of some plant resources and germplasm innovation of Iris L. (Dissertation). Nanjing Agricultural University, China.
Huang, C. X., Liu, F. Y., Gou, Y. N., Deng, J. X., and Li, M. J. (2022). Alternaria vignae sp. nov. (Ascomycota: Pleosporaceae) from Vigna unguiculata in China. Phytotaxa 556, 281–290. doi: 10.11646/phytotaxa.556.3.4
Kahl, S. M., Ulrich, A., Kirichenko, A. A., and Muller, M. E. H. (2015). Phenotypic and phylogenetic segregation of Alternaria infectoria from small-spored Alternaria species isolated from wheat in Germany and Russia. J. Appl. Microbiol. 119, 1637–1650. doi: 10.1111/jam.12951
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lawrence, D. P., Gannibal, P. B., Peever, T. L., and Pryor, B. M. (2013). The sections of Alternaria: formalizing species-group concepts. Mycologia 105, 530–546. doi: 10.3852/12-249
Lawrence, D. P., Park, M. S., and Pryor, B. M. (2012). Nimbya and Embellisia revisited, with nov. comb. for Alternaria celosiae and A. perpunctulata. Mycol. Prog. 11, 799–815. doi: 10.1007/s11557-011-0793-7
Lawrence, D. P., Rotondo, F., and Gannibal, P. B. (2016). Biodiversity and taxonomy of the pleomorphic genus Alternaria. Mycol. Prog. 15, 1–22. doi: 10.1007/s11557-015-1144-x
Li, P., Liu, D., and Yu, B. X. (2020). First report of Alternaria tenuissima causing leaf spot on Iris ensata in Anqing, China. Plant Dis. 104, 1549–1549. doi: 10.1094/PDIS-11-19-2387-PDN
Li, J., Phookamsak, R., Jiang, H., Bhat, D. J., Camporesi, E., Lumyong, S., et al. (2022). Additions to the inventory of the genus Alternaria section Alternaria (Pleosporaceae, Pleosporales) in Italy. J. Fungi 8:898. doi: 10.3390/jof8090898
Liu, Q. L., Li, G. Q., Li, J. Q., and Chen, S. F. (2016). Botrytis eucalypti, a novel species isolated from diseased eucalyptus seedlings in South China. Mycol. Prog. 15, 1057–1079. doi: 10.1007/s11557-016-1229-1
Liu, H. F., Liao, J., Chen, X. Y., Liu, Q. K., and Deng, J. X. (2019). A novel species and a new record of Alternaria isolated from two solanaceae plants in China. Mycol. Prog. 18, 1005–1012. doi: 10.1007/s11557-019-01504-3
Liu, Y. J., Whelen, S., and Hall, B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092
Lu, X. B., Tang, W., Xu, X. H., Sun, H. W., Li, F., Gao, R., et al. (2016). First report of cucumber mosaic virus on Iris tectorum in China. Plant Dis. 100, 1512–1512. doi: 10.1094/PDIS-01-16-0083-PDN
Luo, H., Tao, Y. Q., Fan, X. Y., Oh, S. K., Lu, H. X., and Deng, J. X. (2018). Identification and characterization of Alternaria iridiaustralis causing leaf spot on Iris ensata in China. Mycobiology 46, 168–171. doi: 10.1080/12298093.2018.1454007
Marin-Felix, Y., Hernandez-Restrepo, M., Iturrieta-Gonzalez, I., Garcia, D., Gene, J., Groenewald, J. Z., et al. (2019). Genera of phytopathogenic fungi: GOPHY 3. Stud. Mycol. 94, 1–124. doi: 10.1016/j.simyco.2019.05.001
Meng, S., Torto-Alalibo, T., Chibucos, M. C., Tyler, B. M., and Dean, R. A. (2009). Common processes in pathogenesis by fungal and oomycete plant pathogens, described with Gene ontology terms. BMC Microbiol. 9:S7. doi: 10.1186/1471-2180-9-S1-S7
Meng, F. H., Zhao, R. R., Hu, Z. Y., Wu, B. Q., Lu, H., and Zhao, H.. (2016). Research progress on chemical base and medicinal value of Iris. South. Agric. 10, 91–93. doi: 10.19415/j.cnki.1673-890x.2016.21.054
Nishikawa, J., and Nakashima, C. (2020). Japanese species of Alternaria and their species boundaries based on host range. Fungal Sys. Evol. 5, 197–282. doi: 10.3114/fuse.2020.05.13
Posada, D., and Crandall, K. A. (1998). Model test: testing the model of DNA substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Pryor, B. M., and Gilbertson, R. L. (2000). Molecular phylogenetic relationships amongst Alternaria species and related fungi based upon analysis of nuclear ITS and mt SSU rDNA sequences. Mycol. Res. 104, 1312–1321. doi: 10.1017/S0953756200003002
Rambaut, A., and Drummond, A. (2010). Fig Tree v.1.3.1. Institute of evolutionary biology, University of Edinburgh, Edinburgh, UK.
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Schultes, N. P., Murtishi, B., and Li, D. W. (2017). Phylogenetic relationships of Chlamydomyces, Harzia, Olpitrichum, and their sexual allies, Melanospora and Sphaerodes. Fungal Biol. 121, 890–904. doi: 10.1016/j.funbio.2017.07.004
Simmons, E. G. (2007). Alternaria: An identification manual, CBS Fungal Biodiversity Centre: Utrecht, The Netherlands.
Sofie, L., Michiel, V., Bernard, P., Ayres, D. B., Monica, H., and Kris, A. (2017). Identification of A. arborescens, A. grandis, and A. protenta as new members of the European Alternaria population on potato. Fungal Biology,, 172–188. doi: 10.1016/j.funbio.2016.11.005
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Sun, H., Song, C., Mubeen, I., and Huang, J. (2019). First report of Alternaria tenuissima causing leaf spot disease on Iris tectorum in China. Plant Dis. 103, 1786–1786. doi: 10.1094/PDIS-01-19-0116-PDN
Tahara, S., Ingham, J. L., Hanawa, F., and Mizutani, J. (1991). 1H NMR chemical shift value of the isoflavone 5-hydroxyl proton as a convenient indicator of 6-substitution or 2′-hydroxylation. Phytochemistry 30, 1683–1689. doi: 10.1016/0031-9422(91)84234-J
Thomma, B. P. H. J. (2003). Alternaria spp.: from general saprophyte to specific parasite. Mol. Plant Pathol. 4, 225–236. doi: 10.1046/j.1364-3703.2003.00173.x
Umesha, S., Kong, P., Richardson, P. A., and Hong, C. X. (2010). Bacterial blight of iris caused by pseudomonas syringae in Virginia. Plant Pathol. 57:1176. doi: 10.1111/j.1365-3059.2008.01916.x
Wang, H., Cui, Y. M., and Zhao, C. Q. (2010). Flavonoids of the genus iris (Iridaceae). Mini-Rev. Med. Chem. 10, 643–661. doi: 10.2174/138955710791384027
Wang, C. B., Jiang, N., Zhu, Y. Q., Xue, H., Piao, C. G., and Li, Y. (2022). Colletotrichum truncatum causing anthracnose disease of Iris lactea in Beijing, China. J. Phytopathol. 170, 391–398. doi: 10.1111/jph.13088
Wang, C., Yu, L. Z., Song, S. Y., Li, Y. J., and Tian, Y. M. (2020). Morphological and molecular identification of Aphelenchoides fragariae intercepted in seedlings of Iris bulbs. Plant Prot. 46, 117–121. doi: 10.16688/j.zwbh.2019039
Watanabe, M., Lee, K., Goto, K., Kumagai, S., Sugita-Konishi, Y., and Hara-Kudo, Y. (2010). Rapid and effective DNA extraction method with bead grinding for a large amount of fungal DNA. J. Food Prot. 73, 1077–1084. doi: 10.4315/0362-028X-73.6.1077
White, T.J., Bruns, S., Lee, S., and Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J. and White, T.J. (Eds.) PCR protocols: a guide to methods and applications. Academic Press Inc., New York, 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Woudenberg, J. H. C., Groenewald, J. Z., Binder, M., and Crous, P. W. (2013). Alternaria redefined. Stud. Mycol. 75, 171–212. doi: 10.3114/sim0015
Woudenberg, J. H. C., Seidl, M. F., Groenewald, J. Z., De, V. M., Stielow, J. B., Thomma, B. P. H. J., et al. (2015). Alternaria section Alternaria: species, formae speciales or pathotypes? Stud. Mycol. 82, 1–21. doi: 10.1016/j.simyco.2015.07.001
Woudenberg, J. H. C., Truter, M., Groenewald, J. Z., and Crous, P. W. (2014). Large-spored Alternaria pathogens in section Porri disentangled. Stud. Mycol. 79, 1–47. doi: 10.1016/j.simyco.2014.07.003
Xiang, Q., Duan, F. Y., and Zou, Y. W. (2020). Research progress of Iris in China. Rural Sci. Tech. 11, 50–51. doi: 10.3969/j.issn.1674-7909.2020.36.025
Yang, Z., Meng, T., Bi, X., and Lei, J. (2017). Investigation and evaluation of wild iris resources in Liaoning province, China. Genet. Resour. Crop. Evol. 64, 967–978. doi: 10.1007/s10722-016-0418-8
Yu, S. H. (2015). Fungal Flora of Korea Alternaria and allied genera. National Institute of Biological Resources. Ministry Environ. Korea 1:2.
Zhai, F., Zhang, K., Juan, L. I., et al. (2015). Identification of Alternaria pathogens in Xinxiang area. J. Henan Ins. Sci. Tech.. doi: 10.3969/j.issn.1008-7516.2015.04.007
Zhao, L., Luo, H., Cheng, H., Gou, Y. N., Yu, Z. H., and Deng, J. X. (2022). New species of large-Spored Alternaria in section Porri associated with Compositae plants in China. J. Fungi 8:607. doi: 10.3390/jof8060607
Zhao, Y. T., Noltie, H. J., and Mathew, B. (2000). “Iridaceae,” in Flora of China. eds. Z. Y. Wu and P. H. Raven (Beijing: Science Press), 297–313.
Keywords: Alternaria, Iris, morphology, phylogenetic analysis, pathogenicity
Citation: Gou Y-N, Aung SLL, Htun AA, Huang C-X and Deng J-X (2022) Alternaria species in section Alternaria associated with Iris plants in China. Front. Microbiol. 13:1036950. doi: 10.3389/fmicb.2022.1036950
Edited by:
Nakarin Suwannarach, Chiang Mai University, ThailandReviewed by:
Danushka Sandaruwan Tennakoon, Chiang Mai University, ThailandRungtiwa Phookamsak, Kunming Institute of Botany (CAS), China
Mingkwan Doilom, Zhongkai University of Agriculture and Engineering, China
Copyright © 2022 Gou, Aung, Htun, Huang and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Xin Deng, djxin555@yangtzeu.edu.cn
 Ya-Nan Gou
Ya-Nan Gou Sein Lai Lai Aung1
Sein Lai Lai Aung1 Jian-Xin Deng
Jian-Xin Deng