Lipin-1, a Versatile Regulator of Lipid Homeostasis, Is a Potential Target for Fighting Cancer
Abstract
:1. Introduction
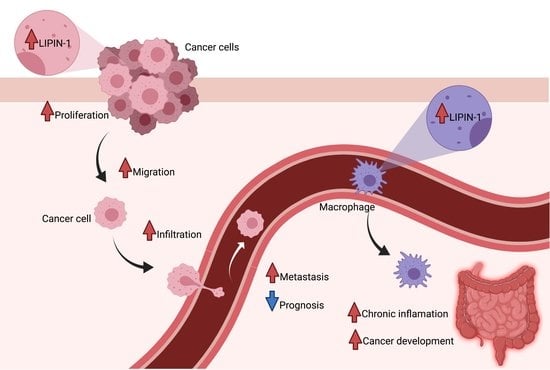
2. Lipin-1 Is Overexpressed in Several Cancer Types and Regulates Cancer Cells Phenotype
3. Mechanisms Regulated by Lipin-1 in Cancer
4. miRNAs as Tumor Suppressors Targeting Lipin-1
5. c-src and mTORC1 Target Lipin-1 to Regulate Cancer Progression
6. The Role of Host Lipin-1 in Inflammation-Driven Cancer Progression
7. Lipin-1 Silencing as a Novel Adjuvant Therapy for Cancer Treatment
8. Propranolol, a Pharmacological Inhibitor of Lipin Enzymatic Activity, Alters Autophagy Homeostasis
9. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ferreira, L.M.; Hebrant, A.; Dumont, J.E. Metabolic reprogramming of the tumor. Oncogene 2012, 31, 3999–4011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ackerman, D.; Simon, M.C. Hypoxia, lipids, and cancer: Surviving the harsh tumor microenvironment. Trends Cell Biol. 2014, 24, 472–478. [Google Scholar] [CrossRef] [Green Version]
- Warburg, O.; Wind, F.; Negelein, E. The Metabolism of Tumors in the Body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Payen, V.L.; Porporato, P.E.; Baselet, B.; Sonveaux, P. Metabolic changes associated with tumor metastasis, part 1: Tumor pH, glycolysis and the pentose phosphate pathway. Cell. Mol. Life Sci. 2016, 73, 1333–1348. [Google Scholar] [CrossRef]
- Porporato, P.E.; Payen, V.L.; Baselet, B.; Sonveaux, P. Metabolic changes associated with tumor metastasis, part 2: Mitochondria, lipid and amino acid metabolism. Cell. Mol. Life Sci. 2016, 73, 1349–1363. [Google Scholar] [CrossRef] [PubMed]
- Snaebjornsson, M.T.; Janaki-Raman, S.; Schulze, A. Greasing the Wheels of the Cancer Machine: The Role of Lipid Metabolism in Cancer. Cell Metab. 2020, 31, 62–76. [Google Scholar] [CrossRef]
- Bensaad, K.; Favaro, E.; Lewis, C.A.; Peck, B.; Lord, S.; Collins, J.M.; Pinnick, K.E.; Wigfield, S.; Buffa, F.M.; Li, J.L.; et al. Fatty acid uptake and lipid storage induced by HIF-1alpha contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 2014, 9, 349–365. [Google Scholar] [CrossRef] [Green Version]
- Sounni, N.E.; Cimino, J.; Blacher, S.; Primac, I.; Truong, A.; Mazzucchelli, G.; Paye, A.; Calligaris, D.; Debois, D.; De Tullio, P.; et al. Blocking lipid synthesis overcomes tumor regrowth and metastasis after antiangiogenic therapy withdrawal. Cell Metab. 2014, 20, 280–294. [Google Scholar] [CrossRef] [Green Version]
- Rohrig, F.; Schulze, A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 2016, 16, 732–749. [Google Scholar] [CrossRef]
- Beckers, A.; Organe, S.; Timmermans, L.; Scheys, K.; Peeters, A.; Brusselmans, K.; Verhoeven, G.; Swinnen, J.V. Chemical inhibition of acetyl-CoA carboxylase induces growth arrest and cytotoxicity selectively in cancer cells. Cancer Res. 2007, 67, 8180–8187. [Google Scholar] [CrossRef] [Green Version]
- De Schrijver, E.; Brusselmans, K.; Heyns, W.; Verhoeven, G.; Swinnen, J.V. RNA interference-mediated silencing of the fatty acid synthase gene attenuates growth and induces morphological changes and apoptosis of LNCaP prostate cancer cells. Cancer Res. 2003, 63, 3799–3804. [Google Scholar]
- Zaidi, N.; Royaux, I.; Swinnen, J.V.; Smans, K. ATP citrate lyase knockdown induces growth arrest and apoptosis through different cell- and environment-dependent mechanisms. Mol. Cancer Ther. 2012, 11, 1925–1935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bacci, M.; Lorito, N.; Smiriglia, A.; Morandi, A. Fat and Furious: Lipid Metabolism in Antitumoral Therapy Response and Resistance. Trends Cancer 2021, 7, 198–213. [Google Scholar] [CrossRef] [PubMed]
- Currie, E.; Schulze, A.; Zechner, R.; Walther, T.C.; Farese, R.V., Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013, 18, 153–161. [Google Scholar] [CrossRef] [Green Version]
- Donkor, J.; Sariahmetoglu, M.; Dewald, J.; Brindley, D.N.; Reue, K. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J. Biol. Chem. 2007, 282, 3450–3457. [Google Scholar] [CrossRef] [Green Version]
- Harris, T.E.; Finck, B.N. Dual function lipin proteins and glycerolipid metabolism. Trends Endocrinol. Metab. 2011, 22, 226–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennedy, E.P. The biological synthesis of phospholipids. Can. J. Biochem. Physiol. 1956, 34, 334–348. [Google Scholar] [CrossRef]
- Csaki, L.S.; Dwyer, J.R.; Fong, L.G.; Tontonoz, P.; Young, S.G.; Reue, K. Lipins, lipinopathies, and the modulation of cellular lipid storage and signaling. Prog. Lipid Res. 2013, 52, 305–316. [Google Scholar] [CrossRef] [Green Version]
- Chae, M.; Jung, J.Y.; Bae, I.H.; Kim, H.J.; Lee, T.R.; Shin, D.W. Lipin-1 expression is critical for keratinocyte differentiation. J. Lipid Res. 2016, 57, 563–573. [Google Scholar] [CrossRef] [Green Version]
- Koh, Y.K.; Lee, M.Y.; Kim, J.W.; Kim, M.; Moon, J.S.; Lee, Y.J.; Ahn, Y.H.; Kim, K.S. Lipin1 is a key factor for the maturation and maintenance of adipocytes in the regulatory network with CCAAT/enhancer-binding protein alpha and peroxisome proliferator-activated receptor gamma 2. J. Biol. Chem. 2008, 283, 34896–34906. [Google Scholar] [CrossRef] [Green Version]
- Chandran, S.; Schilke, R.M.; Blackburn, C.M.R.; Yurochko, A.; Mirza, R.; Scott, R.S.; Finck, B.N.; Woolard, M.D. Lipin-1 Contributes to IL-4 Mediated Macrophage Polarization. Front. Immunol. 2020, 11, 787. [Google Scholar] [CrossRef]
- Meana, C.; Pena, L.; Lorden, G.; Esquinas, E.; Guijas, C.; Valdearcos, M.; Balsinde, J.; Balboa, M.A. Lipin-1 integrates lipid synthesis with proinflammatory responses during TLR activation in macrophages. J. Immunol. 2014, 193, 4614–4622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navratil, A.R.; Vozenilek, A.E.; Cardelli, J.A.; Green, J.M.; Thomas, M.J.; Sorci-Thomas, M.G.; Orr, A.W.; Woolard, M.D. Lipin-1 contributes to modified low-density lipoprotein-elicited macrophage pro-inflammatory responses. Atherosclerosis 2015, 242, 424–432. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Verity, M.A.; Reue, K. Lipin-1 regulates autophagy clearance and intersects with statin drug effects in skeletal muscle. Cell Metab. 2014, 20, 267–279. [Google Scholar] [CrossRef] [Green Version]
- Romani, P.; Brian, I.; Santinon, G.; Pocaterra, A.; Audano, M.; Pedretti, S.; Mathieu, S.; Forcato, M.; Bicciato, S.; Manneville, J.B.; et al. Extracellular matrix mechanical cues regulate lipid metabolism through Lipin-1 and SREBP. Nat. Cell Biol. 2019, 21, 338–347. [Google Scholar] [CrossRef] [Green Version]
- Finck, B.N.; Gropler, M.C.; Chen, Z.; Leone, T.C.; Croce, M.A.; Harris, T.E.; Lawrence, J.C., Jr.; Kelly, D.P. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 2006, 4, 199–210. [Google Scholar] [CrossRef] [Green Version]
- Donkor, J.; Zhang, P.; Wong, S.; O’Loughlin, L.; Dewald, J.; Kok, B.P.; Brindley, D.N.; Reue, K. A conserved serine residue is required for the phosphatidate phosphatase activity but not the transcriptional coactivator functions of lipin-1 and lipin-2. J. Biol. Chem. 2009, 284, 29968–29978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.B.; Kumar, A.; Wang, L.; Liu, G.H.; Keller, S.R.; Lawrence, J.C., Jr.; Finck, B.N.; Harris, T.E. Lipin 1 represses NFATc4 transcriptional activity in adipocytes to inhibit secretion of inflammatory factors. Mol. Cell Biol. 2010, 30, 3126–3139. [Google Scholar] [CrossRef] [Green Version]
- Peterson, T.R.; Sengupta, S.S.; Harris, T.E.; Carmack, A.E.; Kang, S.A.; Balderas, E.; Guertin, D.A.; Madden, K.L.; Carpenter, A.E.; Finck, B.N.; et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 2011, 146, 408–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phan, J.; Peterfy, M.; Reue, K. Lipin expression preceding peroxisome proliferator-activated receptor-gamma is critical for adipogenesis in vivo and in vitro. J. Biol. Chem. 2004, 279, 29558–29564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, Y.; Kwon, S.; Ham, S.; Lee, D.; Park, H.H.; Yamaoka, Y.; Jeong, D.E.; Artan, M.; Altintas, O.; Park, S.; et al. Caenorhabditis elegans Lipin 1 moderates the lifespan-shortening effects of dietary glucose by maintaining omega-6 polyunsaturated fatty acids. Aging Cell 2020, 19, e13150. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, M. Diverse roles of phosphatidate phosphatases in insect development and metabolism. Insect Biochem. Mol. Biol. 2020, 103469. [Google Scholar] [CrossRef]
- Schmitt, S.; Ugrankar, R.; Greene, S.E.; Prajapati, M.; Lehmann, M. Drosophila Lipin interacts with insulin and TOR signaling pathways in the control of growth and lipid metabolism. J. Cell Sci. 2015, 128, 4395–4406. [Google Scholar] [CrossRef] [Green Version]
- Lutkewitte, A.J.; Finck, B.N. Regulation of Signaling and Metabolism by Lipin-mediated Phosphatidic Acid Phosphohydrolase Activity. Biomolecules 2020, 10, 1386. [Google Scholar] [CrossRef] [PubMed]
- Reue, K.; Wang, H. Mammalian lipin phosphatidic acid phosphatases in lipid synthesis and beyond: Metabolic and inflammatory disorders. J. Lipid Res. 2019, 60, 728–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brohee, L.; Demine, S.; Willems, J.; Arnould, T.; Colige, A.C.; Deroanne, C.F. Lipin-1 regulates cancer cell phenotype and is a potential target to potentiate rapamycin treatment. Oncotarget 2015, 6, 11264–11280. [Google Scholar] [CrossRef]
- Fan, X.; Weng, Y.; Bai, Y.; Wang, Z.; Wang, S.; Zhu, J.; Zhang, F. Lipin-1 determines lung cancer cell survival and chemotherapy sensitivity by regulation of endoplasmic reticulum homeostasis and autophagy. Cancer Med. 2018, 7, 2541–2554. [Google Scholar] [CrossRef]
- He, J.; Zhang, F.; Tay, L.W.R.; Boroda, S.; Nian, W.; Levental, K.R.; Levental, I.; Harris, T.E.; Chang, J.T.; Du, G. Lipin-1 regulation of phospholipid synthesis maintains endoplasmic reticulum homeostasis and is critical for triple-negative breast cancer cell survival. FASEB J. 2017, 31, 2893–2904. [Google Scholar] [CrossRef] [Green Version]
- Ingram, L.M.; Finnerty, M.C.; Mansoura, M.; Chou, C.W.; Cummings, B.S. Identification of lipidomic profiles associated with drug-resistant prostate cancer cells. Lipids Health Dis. 2021, 20, 15. [Google Scholar] [CrossRef]
- Santuario-Facio, S.K.; Cardona-Huerta, S.; Perez-Paramo, Y.X.; Trevino, V.; Hernandez-Cabrera, F.; Rojas-Martinez, A.; Uscanga-Perales, G.; Martinez-Rodriguez, J.L.; Martinez-Jacobo, L.; Padilla-Rivas, G.; et al. A New Gene Expression Signature for Triple Negative Breast Cancer Using Frozen Fresh Tissue before Neoadjuvant Chemotherapy. Mol. Med. 2017, 23, 101–111. [Google Scholar] [CrossRef]
- Dinarvand, N.; Khanahmad, H.; Hakimian, S.M.; Sheikhi, A.; Rashidi, B.; Bakhtiari, H.; Pourfarzam, M. Expression and clinicopathological significance of lipin-1 in human breast cancer and its association with p53 tumor suppressor gene. J. Cell. Physiol. 2020, 235, 5835–5846. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kim, G.; Lim, S.C.; Choi, H.S. LPIN1 promotes epithelial cell transformation and mammary tumourigenesis via enhancing insulin receptor substrate 1 stability. Carcinogenesis 2016, 37, 1199–1209. [Google Scholar] [CrossRef]
- Zhao, S.; Li, J.; Zhang, G.; Wang, Q.; Wu, C.; Zhang, Q.; Wang, H.; Sun, P.; Xiang, R.; Yang, S. Exosomal miR-451a Functions as a Tumor Suppressor in Hepatocellular Carcinoma by Targeting LPIN1. Cell Physiol. Biochem. 2019, 53, 19–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Ma, H.L. MiRNA-584 suppresses the progression of ovarian cancer by negatively regulating LPIN1. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Liu, Z.; Hu, H.H.; Yang, Y.; Li, T.Y.; Lin, Z.Z.; Ye, J.; Chen, J.; Huang, X.; Liu, D.T.; et al. Proto-oncogene Src links lipogenesis via lipin-1 to breast cancer malignancy. Nat. Commun. 2020, 11, 5842. [Google Scholar] [CrossRef] [PubMed]
- MacVicar, T.; Ohba, Y.; Nolte, H.; Mayer, F.C.; Tatsuta, T.; Sprenger, H.G.; Lindner, B.; Zhao, Y.; Li, J.; Bruns, C.; et al. Lipid signalling drives proteolytic rewiring of mitochondria by YME1L. Nature 2019, 575, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Schilke, R.M.; Blackburn, C.M.R.; Rao, S.; Krzywanski, D.M.; Finck, B.N.; Woolard, M.D. Macrophage-Associated Lipin-1 Promotes beta-Oxidation in Response to Proresolving Stimuli. Immunohorizons 2020, 4, 659–669. [Google Scholar] [CrossRef]
- Hou, J.; Karin, M.; Sun, B. Targeting cancer-promoting inflammation-have anti-inflammatory therapies come of age? Nat. Rev. Clin. Oncol. 2021. [Google Scholar] [CrossRef]
- Wang, K.; Karin, M. Tumor-Elicited Inflammation and Colorectal Cancer. Adv. Cancer Res. 2015, 128, 173–196. [Google Scholar] [CrossRef]
- Vozenilek, A.E.; Navratil, A.R.; Green, J.M.; Coleman, D.T.; Blackburn, C.M.R.; Finney, A.C.; Pearson, B.H.; Chrast, R.; Finck, B.N.; Klein, R.L.; et al. Macrophage-Associated Lipin-1 Enzymatic Activity Contributes to Modified Low-Density Lipoprotein-Induced Proinflammatory Signaling and Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 324–334. [Google Scholar] [CrossRef] [Green Version]
- Meana, C.; Garcia-Rostan, G.; Pena, L.; Lorden, G.; Cubero, A.; Orduna, A.; Gyorffy, B.; Balsinde, J.; Balboa, M.A. The phosphatidic acid phosphatase lipin-1 facilitates inflammation-driven colon carcinogenesis. JCI Insight 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Kwitkowski, V.E.; Prowell, T.M.; Ibrahim, A.; Farrell, A.T.; Justice, R.; Mitchell, S.S.; Sridhara, R.; Pazdur, R. FDA approval summary: Temsirolimus as treatment for advanced renal cell carcinoma. Oncologist 2010, 15, 428–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motzer, R.J.; Escudier, B.; Oudard, S.; Hutson, T.E.; Porta, C.; Bracarda, S.; Grunwald, V.; Thompson, J.A.; Figlin, R.A.; Hollaender, N.; et al. Efficacy of everolimus in advanced renal cell carcinoma: A double-blind, randomised, placebo-controlled phase III trial. Lancet 2008, 372, 449–456. [Google Scholar] [CrossRef]
- Fang, Z.; Zhang, T.; Dizeyi, N.; Chen, S.; Wang, H.; Swanson, K.D.; Cai, C.; Balk, S.P.; Yuan, X. Androgen Receptor Enhances p27 Degradation in Prostate Cancer Cells through Rapid and Selective TORC2 Activation. J. Biol. Chem. 2012, 287, 2090–2098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saijo, K.; Imai, H.; Chikamatsu, S.; Narita, K.; Katoh, T.; Ishioka, C. Antitumor activity and pharmacologic characterization of the depsipeptide analog as a novel histone deacetylase/ phosphatidylinositol 3-kinase dual inhibitor. Cancer Sci. 2017, 108, 1469–1475. [Google Scholar] [CrossRef] [Green Version]
- Imai, H.; Saijo, K.; Chikamatsu, S.; Kawamura, Y.; Ishioka, C. LPIN1 downregulation enhances anticancer activity of the novel HDAC/PI3K dual inhibitor FK-A11. Cancer Sci. 2021, 112, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Thorburn, A.; Thamm, D.H.; Gustafson, D.L. Autophagy and cancer therapy. Mol. Pharmacol. 2014, 85, 830–838. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J. Targeting mTOR by CZ415 Suppresses Cell Proliferation and Promotes Apoptosis via Lipin-1 in Cervical Cancer In Vitro and In Vivo. Reprod. Sci. 2021, 28, 524–531. [Google Scholar] [CrossRef]
- Byington, R.P. Beta-blocker heart attack trial: Design, methods, and baseline results. Beta-blocker heart attack trial research group. Control Clin. Trials 1984, 5, 382–437. [Google Scholar] [CrossRef]
- Stiles, G.L.; Caron, M.G.; Lefkowitz, R.J. Beta-adrenergic receptors: Biochemical mechanisms of physiological regulation. Physiol. Rev. 1984, 64, 661–743. [Google Scholar] [CrossRef]
- Jamal, Z.; Martin, A.; Gomez-Munoz, A.; Brindley, D.N. Plasma membrane fractions from rat liver contain a phosphatidate phosphohydrolase distinct from that in the endoplasmic reticulum and cytosol. J. Biol. Chem. 1991, 266, 2988–2996. [Google Scholar] [CrossRef]
- Koul, O.; Hauser, G. Modulation of rat brain cytosolic phosphatidate phosphohydrolase: Effect of cationic amphiphilic drugs and divalent cations. Arch. Biochem. Biophys. 1987, 253, 453–461. [Google Scholar] [CrossRef]
- Pantziarka, P.; Bouche, G.; Sukhatme, V.; Meheus, L.; Rooman, I.; Sukhatme, V.P. Repurposing Drugs in Oncology (ReDO)-Propranolol as an anti-cancer agent. Ecancermedicalscience 2016, 10, 680. [Google Scholar] [CrossRef] [Green Version]
- Pantziarka, P.; Bryan, B.A.; Crispino, S.; Dickerson, E.B. Propranolol and breast cancer-a work in progress. Ecancermedicalscience 2018, 12, ed82. [Google Scholar] [CrossRef]
- Chaudhary, K.R.; Yan, S.X.; Heilbroner, S.P.; Sonett, J.R.; Stoopler, M.B.; Shu, C.; Halmos, B.; Wang, T.J.C.; Hei, T.K.; Cheng, S.K. Effects of beta-Adrenergic Antagonists on Chemoradiation Therapy for Locally Advanced Non-Small Cell Lung Cancer. J. Clin. Med. 2019, 8, 575. [Google Scholar] [CrossRef] [Green Version]
- Pasquier, E.; Andre, N.; Street, J.; Chougule, A.; Rekhi, B.; Ghosh, J.; Philip, D.S.J.; Meurer, M.; MacKenzie, K.L.; Kavallaris, M.; et al. Effective Management of Advanced Angiosarcoma by the Synergistic Combination of Propranolol and Vinblastine-based Metronomic Chemotherapy: A Bench to Bedside Study. EBioMedicine 2016, 6, 87–95. [Google Scholar] [CrossRef] [Green Version]
- Rico, M.; Baglioni, M.; Bondarenko, M.; Laluce, N.C.; Rozados, V.; Andre, N.; Carre, M.; Scharovsky, O.G.; Menacho Marquez, M. Metformin and propranolol combination prevents cancer progression and metastasis in different breast cancer models. Oncotarget 2017, 8, 2874–2889. [Google Scholar] [CrossRef] [Green Version]
- Saha, J.; Kim, J.H.; Amaya, C.N.; Witcher, C.; Khammanivong, A.; Korpela, D.M.; Brown, D.R.; Taylor, J.; Bryan, B.A.; Dickerson, E.B. Propranolol Sensitizes Vascular Sarcoma Cells to Doxorubicin by Altering Lysosomal Drug Sequestration and Drug Efflux. Front. Oncol. 2020, 10, 614288. [Google Scholar] [CrossRef]
- Farah, B.L.; Sinha, R.A.; Wu, Y.; Singh, B.K.; Zhou, J.; Bay, B.H.; Yen, P.M. beta-Adrenergic agonist and antagonist regulation of autophagy in HepG2 cells, primary mouse hepatocytes, and mouse liver. PLoS ONE 2014, 9, e98155. [Google Scholar] [CrossRef] [Green Version]
- Schonthal, A.H. Pharmacological targeting of endoplasmic reticulum stress signaling in cancer. Biochem. Pharmacol. 2013, 85, 653–666. [Google Scholar] [CrossRef] [PubMed]
- Brohee, L.; Peulen, O.; Nusgens, B.; Castronovo, V.; Thiry, M.; Colige, A.C.; Deroanne, C.F. Propranolol sensitizes prostate cancer cells to glucose metabolism inhibition and prevents cancer progression. Sci. Rep. 2018, 8, 7050. [Google Scholar] [CrossRef]
- Lucido, C.T.; Miskimins, W.K.; Vermeer, P.D. Propranolol Promotes Glucose Dependence and Synergizes with Dichloroacetate for Anti-Cancer Activity in HNSCC. Cancers 2018, 10, 476. [Google Scholar] [CrossRef] [Green Version]
- Farrow, J.M.; Yang, J.C.; Evans, C.P. Autophagy as a modulator and target in prostate cancer. Nat. Rev. Urol. 2014, 11, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Peterfy, M.; Phan, J.; Reue, K. Alternatively spliced lipin isoforms exhibit distinct expression pattern, subcellular localization, and role in adipogenesis. J. Biol. Chem. 2005, 280, 32883–32889. [Google Scholar] [CrossRef] [Green Version]
- Han, G.S.; Carman, G.M. Characterization of the human LPIN1-encoded phosphatidate phosphatase isoforms. J. Biol. Chem. 2010, 285, 14628–14638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Csaki, L.S.; Reue, K. Lipins: Multifunctional lipid metabolism proteins. Annu. Rev. Nutr. 2010, 30, 257–272. [Google Scholar] [CrossRef] [Green Version]
- Bi, L.; Jiang, Z.; Zhou, J. The role of lipin-1 in the pathogenesis of alcoholic fatty liver. Alcohol Alcohol. 2015, 50, 146–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Cheng, Y.; Wu, W.; Liu, Y.; Wei, N.; Feng, X.; Xie, Z.; Feng, Y. SRSF10 regulates alternative splicing and is required for adipocyte differentiation. Mol. Cell. Biol. 2014, 34, 2198–2207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manmontri, B.; Sariahmetoglu, M.; Donkor, J.; Bou Khalil, M.; Sundaram, M.; Yao, Z.; Reue, K.; Lehner, R.; Brindley, D.N. Glucocorticoids and cyclic AMP selectively increase hepatic lipin-1 expression, and insulin acts antagonistically. J. Lipid Res. 2008, 49, 1056–1067. [Google Scholar] [CrossRef] [Green Version]
- Gropler, M.C.; Harris, T.E.; Hall, A.M.; Wolins, N.E.; Gross, R.W.; Han, X.; Chen, Z.; Finck, B.N. Lipin 2 is a liver-enriched phosphatidate phosphohydrolase enzyme that is dynamically regulated by fasting and obesity in mice. J. Biol. Chem. 2009, 284, 6763–6772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, D.; Seo, W.Y.; Yoon, Y.S.; Kim, Y.N.; Kim, S.S.; Kim, H.J.; Park, T.S.; Choi, C.S.; Koo, S.H. Endoplasmic reticulum stress promotes LIPIN2-dependent hepatic insulin resistance. Diabetes 2011, 60, 1072–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balsinde, J.; Dennis, E.A. Bromoenol lactone inhibits magnesium-dependent phosphatidate phosphohydrolase and blocks triacylglycerol biosynthesis in mouse P388D1 macrophages. J. Biol. Chem. 1996, 271, 31937–31941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khayyo, V.I.; Hoffmann, R.M.; Wang, H.; Bell, J.A.; Burke, J.E.; Reue, K.; Airola, M.V. Crystal structure of a lipin/Pah phosphatidic acid phosphatase. Nat. Commun. 2020, 11, 1309. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brohée, L.; Crémer, J.; Colige, A.; Deroanne, C. Lipin-1, a Versatile Regulator of Lipid Homeostasis, Is a Potential Target for Fighting Cancer. Int. J. Mol. Sci. 2021, 22, 4419. https://doi.org/10.3390/ijms22094419
Brohée L, Crémer J, Colige A, Deroanne C. Lipin-1, a Versatile Regulator of Lipid Homeostasis, Is a Potential Target for Fighting Cancer. International Journal of Molecular Sciences. 2021; 22(9):4419. https://doi.org/10.3390/ijms22094419
Chicago/Turabian StyleBrohée, Laura, Julie Crémer, Alain Colige, and Christophe Deroanne. 2021. "Lipin-1, a Versatile Regulator of Lipid Homeostasis, Is a Potential Target for Fighting Cancer" International Journal of Molecular Sciences 22, no. 9: 4419. https://doi.org/10.3390/ijms22094419