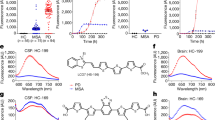
Abstract
Protein misfolding and aggregation are characteristic of a wide range of neurodegenerative disorders, including Alzheimer and Parkinson diseases. A hallmark of these diseases is the aggregation of otherwise soluble and functional proteins into amyloid aggregates. Although for many decades such amyloid deposits have been thought to be responsible for disease progression, it is now increasingly recognized that the misfolded protein oligomers formed during aggregation are, instead, the main agents causing pathological processes. These oligomers are transient and heterogeneous, which makes it difficult to detect and quantify them, generating confusion about their exact role in disease. The lack of suitable methods to address these challenges has hampered efforts to investigate the molecular mechanisms of oligomer toxicity and to develop oligomer-based diagnostic and therapeutic tools to combat protein misfolding diseases. In this Review, we describe methods to quantify misfolded protein oligomers, with particular emphasis on diagnostic applications as disease biomarkers and on therapeutic applications as target biomarkers. The development of these methods is ongoing, and we discuss the challenges that remain to be addressed to establish measurement tools capable of overcoming existing limitations and to meet present needs.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Alzheimer’s Disease International. World Alzheimer report 2019: attitudes to dementia. Alzheimer’s Disease International https://www.alzint.org/resource/world-alzheimer-report-2019/ (2019).
Cummings, J., Lee, G., Ritter, A., Sabbagh, M. & Zhong, K. Alzheimer’s disease drug development pipeline: 2019. Alzheimers Dement. 5, 272–293 (2019).
Cummings, J. L. Translational scoring of candidate treatments for Alzheimer’s disease: a systematic approach. Dement. Geriatr. Cogn. Disord. 49, 22–37 (2020).
Hampel, H. et al. Biomarkers for Alzheimer’s disease: academic, industry and regulatory perspectives. Nat. Rev. Drug Discov. 9, 560–574 (2010).
Aisen, P. S. et al. The future of anti-amyloid trials. J. Prev. Alzheimers Dis. 7, 146–151 (2020).
U.S. Department of Health and Human Services, Food and Drug Administration, Center for Drug Evaluation and Research (CDER) and Center for Biologics Evaluation and Research (CBER). Early Alzheimer’s disease: developing drugs for treatment (2018).
Jack, C. R. et al. NIA-AA research framework: toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 14, 535–562 (2018).
Hardy, J. & Selkoe, D. J. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science 297, 353–356 (2002).
Selkoe, D. J. & Hardy, J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 8, 595–608 (2016).
Spillantini, M. G. et al. α-Synuclein in Lewy bodies. Nature 338, 839–840 (1997).
Braak, H. & Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 (1991).
Prots, I. et al. α-Synuclein oligomers induce early axonal dysfunction in human iPSC-based models of synucleinopathies. Proc. Natl Acad. Sci. USA 115, 7813–7818 (2018).
Haass, C. & Selkoe, D. J. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid β-peptide. Nat. Rev. Mol. Cell Biol. 8, 101–112 (2007).
Benilova, I., Karran, E. & De Strooper, B. The toxic Aβ oligomer and Alzheimer’s disease: an emperor in need of clothes. Nat. Neurosci. 15, 349–357 (2012).
Flagmeier, P. et al. Ultrasensitive measurement of Ca2+ influx into lipid vesicles induced by protein aggregates. Angew. Chem. Int. Ed. 56, 7750–7754 (2017). This article reports a high-throughput oligomer quantification assay based on the measurement of Ca2+ entry into individual lipid vesicles.
Fusco, G. et al. Structural basis of membrane disruption and cellular toxicity by α-synuclein oligomers. Science 358, 1440–1443 (2017).
Cremades, N. et al. Direct observation of the interconversion of normal and toxic forms of α-synuclein. Cell 149, 1048–1059 (2012).
Tucker, S. et al. The murine version of BAN2401 (mAb158) selectively reduces amyloid-β protofibrils in brain and cerebrospinal fluid of tg-ArcSwe mice. J. Alzheimers Dis. 43, 575–588 (2015). This work describes accurate enzyme-linked immunosorbent assay (ELISA) measurements of amyloid-β protofibrils in mouse brain and cerebrospinal fluid.
De, S. et al. Soluble aggregates present in cerebrospinal fluid change in size and mechanism of toxicity during Alzheimer’s disease progression. Acta Neuropathol. Commun. 7, 120 (2019).
Hong, W. et al. Diffusible, highly bioactive oligomers represent a critical minority of soluble Aβ in Alzheimer’s disease brain. Acta Neuropathol. 136, 19–40 (2018). This article describes characterization of the highly heterogeneous nature of misfolded protein oligomers.
Walsh, D. M. & Selkoe, D. J. Amyloid β-protein and beyond: the path forward in Alzheimer’s disease. Curr. Opin. Neurobiol. 61, 116–124 (2020).
Majbour, N. K. et al. Oligomeric and phosphorylated alpha-synuclein as potential CSF biomarkers for Parkinson’s disease. Mol. Neurodegener. 11, 7 (2016).
Saijo, E. et al. 4-Repeat tau seeds and templating subtypes as brain and CSF biomarkers of frontotemporal lobar degeneration. Acta Neuropathol. 139, 63–77 (2020).
Hölttä, M. et al. Evaluating amyloid-β oligomers in cerebrospinal fluid as a biomarker for Alzheimer’s disease. PLoS ONE 8, e66381 (2013). One of the first reports on the use of amyloid-β oligomers as diagnostic markers for Alzheimer disease.
Wang-Dietrich, L. et al. The amyloid-β oligomer count in cerebrospinal fluid is a biomarker for Alzheimer’s disease. J. Alzheimers Dis. 34, 985–994 (2013).
Yang, T. et al. Target engagement in an alzheimer trial: crenezumab lowers amyloid β oligomers in cerebrospinal fluid. Ann. Neurol. 86, 215–224 (2019). This paper demonstrates the use of an Erenna immunoassay to show target engagement of an antibody to amyloid-β aggregation in clinical trials.
Yang, T. et al. A highly sensitive novel immunoassay specifically detects low levels of soluble Aβ oligomers in human cerebrospinal fluid. Alzheimers Res. Ther. 7, 14 (2015).
Georganopoulou, D. G. et al. Nanoparticle-based detection in cerebral spinal fluid of a soluble pathogenic biomarker for Alzheimer’s disease. Proc. Natl Acad. Sci. USA 102, 2273–2276 (2005).
Savage, M. J. et al. A sensitive Aβ oligomer assay discriminates Alzheimer’s and aged control cerebrospinal fluid. J. Neurosci. 34, 2884–2897 (2014).
Dujardin, S. et al. Tau molecular diversity contributes to clinical heterogeneity in Alzheimer’s disease. Nat. Med. 26, 1256–1263 (2020).
Wang, Z. et al. Human brain-derived Aβ oligomers bind to synapses and disrupt synaptic activity in a manner that requires APP. J. Neurosci. 37, 11947–11966 (2017).
Walsh, D. M. et al. Naturally secreted oligomers of amyloid β protein potently inhibit hippocampal long-term potentiation in vivo. Nature 416, 535–539 (2002).
Wang, H.-W. et al. Soluble oligomers of β amyloid (1-42) inhibit long-term potentiation but not long-term depression in rat dentate gyrus. Brain Res. 924, 133–140 (2002).
Li, S. & Selkoe, D. J. A mechanistic hypothesis for the impairment of synaptic plasticity by soluble Aβ oligomers from Alzheimer’s brain. J. Neurochem. 154, 583–597 (2020).
Lesné, S. E. et al. Brain amyloid-β oligomers in ageing and Alzheimer’s disease. Brain 136, 1383–1398 (2013).
Brinkmalm, G. et al. Identification of neurotoxic cross-linked amyloid-β dimers in the Alzheimer’s brain. Brain 142, 1441–1457 (2019).
Wang, Y. & Mandelkow, E. Tau in physiology and pathology. Nat. Rev. Neurosci. 17, 22–35 (2015).
Kopeikina, K. J., Hyman, B. J. & Spires-Jones, T. L. Soluble forms of tau are toxic in Alzheimer’s disease. Transl Neurosci. 3, 223–233 (2012).
Kaniyappan, S., Chandupatla, R. R., Mandelkow, E. M. & Mandelkow, E. Extracellular low-n oligomers of tau cause selective synaptotoxicity without affecting cell viability. Alzheimers Dement. 13, 1270–1291 (2017).
Hill, E., Karikari, T. K., Moffat, K. G., Richardson, M. J. E. & Wall, M. J. Introduction of tau oligomers into cortical neurons alters action potential dynamics and disrupts synaptic transmission and plasticity. eNeuro 6, 5 (2019).
Fá, M. et al. Extracellular tau oligomers produce an immediate impairment of LTP and memory. Sci. Rep. 6, 19393 (2016).
Lasagna-Reeves, C. A. et al. Alzheimer brain-derived tau oligomers propagate pathology from endogenous tau. Sci. Rep. 2, 700 (2012).
Wu, J. W. et al. Small misfolded tau species are internalized via bulk endocytosis and anterogradely and retrogradely transported in neurons. J. Biol. Chem. 288, 1856–1870 (2013).
Das, R., Balmik, A. A. & Chinnathambi, S. Phagocytosis of full-length tau oligomers by actin-remodeling of activated microglia. J. Neuroinflammation 17, 10 (2020).
Clavaguera, F. et al. Transmission and spreading of tauopathy in transgenic mouse brain. Nat. Cell Biol. 11, 909–913 (2009).
Jiang, L. et al. TIA1 regulates the generation and response to toxic tau oligomers. Acta Neuropathol. 137, 259–277 (2019).
Ingelsson, M. Alpha-synuclein oligomers — neurotoxic molecules in Parkinson’s disease and other Lewy body disorders. Front. Neurosci. 10, 408 (2016).
Bengoa-Vergniory, N., Roberts, R. F., Wade-Martins, R. & Alegre-Abarrategui, J. Alpha-synuclein oligomers: a new hope. Acta Neuropathol. 134, 819–838 (2017).
Alam, P., Bousset, L., Melki, R. & Otzen, D. E. α-Synuclein oligomers and fibrils: a spectrum of species, a spectrum of toxicities. J. Neurochem. 150, 522–534 (2019).
Kayed, R., Dettmer, U. & Lesné, S. E. Soluble endogenous oligomeric α-synuclein species in neurodegenerative diseases: expression, spreading, and cross-talk. J. Parkinsons Dis. 10, 791–818 (2020).
Winner, B. et al. In vivo demonstration that α-synuclein oligomers are toxic. Proc. Natl Acad. Sci. USA 108, 4194–4199 (2011).
van Diggelen, F. et al. Two conformationally distinct α-synuclein oligomers share common epitopes and the ability to impair long-term potentiation. PLoS ONE 14, e0213663 (2019).
Di Maio, R. et al. α-Synuclein binds to TOM20 and inhibits mitochondrial protein import in Parkinson’s disease. Sci. Transl Med. 8, 342ra78 (2016).
Roberts, R. F., Wade-Martins, R. & Alegre-Abarrategui, J. Direct visualization of alpha-synuclein oligomers reveals previously undetected pathology in Parkinson’s disease brain. Brain 138, 1642–1657 (2015).
Danzer, K. M. et al. Exosomal cell-to-cell transmission of alpha synuclein oligomers. Mol. Neurodegener. 7, 42 (2012).
Knowles, T. P. J. et al. An analytical solution to the kinetics of breakable filament assembly. Science 326, 1533–1537 (2009).
Cohen, S. I. A. et al. Proliferation of amyloid-42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl Acad. Sci. USA 110, 9758–9763 (2013).
Habchi, J. et al. Cholesterol catalyses Aβ42 aggregation through a heterogeneous nucleation pathway in the presence of lipid membranes. Nat. Chem. 10, 673–683 (2018).
Michaels, T. C. T. et al. Dynamics of oligomer populations formed during the aggregation of Alzheimer’s Aβ42 peptide. Nat. Chem. 12, 445–451 (2020). This work shows that Aβ42 oligomers should undergo a structural conversion step after their initial formation in order to grow into amyloid fibrils.
Koffie, R. M. et al. Oligomeric amyloid β associates with postsynaptic densities and correlates with excitatory synapse loss near senile plaques. Proc. Natl Acad. Sci. USA 106, 4012–4017 (2009).
Galvagnion, C. et al. Lipid vesicles trigger α-synuclein aggregation by stimulating primary nucleation. Nat. Chem. Biol. 11, 229–234 (2015).
Buell, A. K. et al. Solution conditions determine the relative importance of nucleation and growth processes in α-synuclein aggregation. Proc. Natl Acad. Sci. USA 111, 7671–7676 (2014).
Shammas, S. L. et al. A mechanistic model of tau amyloid aggregation based on direct observation of oligomers. Nat. Commun. 6, 7025 (2015).
Dear, A. J. et al. Kinetic diversity of amyloid oligomers. Proc. Natl Acad. Sci. USA 117, 12087–12094 (2020).
Lambert, M. P. et al. Diffusible, nonfibrillar ligands derived from Aβ1–42 are potent central nervous system neurotoxins. Proc. Natl Acad. Sci. USA 95, 6448–6453 (1998).
Jackson, S. J. et al. Short fibrils constitute the major species of seed-competent tau in the brains of mice transgenic for human p301s tau. J. Neurosci. 36, 762–772 (2016).
Knowles, T. P. J., Vendruscolo, M. & Dobson, C. M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 15, 384–396 (2014).
Kayed, R. et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300, 486–489 (2003).
Lassen, L. B. et al. ELISA method to detect α-synuclein oligomers in cell and animal models. PLoS ONE 13, e0196056 (2018).
Wang, M. J. et al. Oligomeric forms of amyloid-β protein in plasma as a potential blood-based biomarker for Alzheimer’s disease. Alzheimers Res. Ther. 9, 98 (2017).
Youn, Y. C. et al. Blood amyloid-β oligomerization associated with neurodegeneration of Alzheimer’s disease. Alzheimers Res. Ther. 11, 40 (2019).
El-Agnaf, O. M. A. et al. Detection of oligomeric forms of α-synuclein protein in human plasma as a potential biomarker for Parkinson’s disease. FASEB J. 20, 419–425 (2006).
Bruggink, K. A. et al. Amyloid-β oligomer detection by ELISA in cerebrospinal fluid and brain tissue. Anal. Biochem. 433, 112–120 (2013).
Pryor, N. E., Moss, M. A. & Hestekin, C. N. Unraveling the early events of amyloid-β protein (Aβ) aggregation: techniques for the determination of Aβ aggregate size. Int. J. Mol. Sci. 13, 3038–3072 (2012).
Sharon, R. et al. The formation of highly soluble oligomers of α-synuclein is regulated by fatty acids and enhanced in Parkinson’s disease. Neuron 37, 583–595 (2003).
Sengupta, U. et al. Tau oligomers in cerebrospinal fluid in Alzheimer’s disease. Ann. Clin. Transl Neurol. 4, 226–235 (2017).
Kuhle, J. et al. Comparison of three analytical platforms for quantification of the neurofilament light chain in blood samples: ELISA, electrochemiluminescence immunoassay and Simoa. Clin. Chem. Lab. Med. 54, 1655–1661 (2016).
Rissin, D. M. & Walt, D. R. Digital readout of target binding with attomole detection limits via enzyme amplification in femtoliter arrays. J. Am. Chem. Soc. 128, 6286–6287 (2006).
Rissin, D. M. & Walt, D. R. Digital concentration readout of single enzyme molecules using femtoliter arrays and Poisson statistics. Nano Lett. 6, 520–523 (2006).
Hwang, S. S. et al. Detection of amyloid β oligomers toward early diagnosis of Alzheimer’s disease. Anal. Biochem. 566, 40–45 (2019). This work compares the quartz crystal microbalance (QCM) and single-molecule array (SIMOA) assay for the quantification of misfolded protein oligomers.
Li, D. & Mielke, M. M. An update on blood-based markers of Alzheimer’s disease using the SiMoA platform. Neurol. Ther. 8, 73–82 (2019).
Wu, C., Garden, P. M. & Walt, D. R. Ultrasensitive detection of attomolar protein concentrations by dropcast single molecule assays. J. Am. Chem. Soc. 142, 12314–12323 (2020).
Fulton, R. J., McDade, R. L., Smith, P. L., Kienker, L. J. & Kettman, J. R. Advanced multiplexed analysis with the FlowMetrixTM system. Clin. Chem. 43, 1749–1756 (1997).
Herskovits, A. Z., Locascio, J. J., Peskind, E. R., Li, G. & Hyman, B. T. A Luminex assay detects amyloid β oligomers in Alzheimer’s disease cerebrospinal fluid. PLoS ONE 8, e67898 (2013).
Fukumoto, H. et al. High-molecular-weight β-amyloid oligomers are elevated in cerebrospinal fluid of Alzheimer patients. FASEB J. 24, 2716–2726 (2010).
Esparza, T. J. et al. Amyloid-beta oligomerization in Alzheimer dementia versus high-pathology controls. Ann. Neurol. 73, 104–119 (2013).
Park, M. C. et al. Droplet-based magnetic bead immunoassay using microchannel-connected multiwell plates (μCHAMPs) for the detection of amyloid beta oligomers. Lab Chip 16, 2245–2253 (2016).
Takahashi, T. & Mihara, H. FRET detection of amyloid β-peptide oligomerization using a fluorescent protein probe presenting a pseudo-amyloid structure. Chem. Commun. 48, 1568–1570 (2012).
Ferreon, A. C. M., Gambin, Y., Lemke, E. A. & Deniz, A. A. Interplay of α-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc. Natl Acad. Sci. USA 106, 5645–5650 (2009).
Tosatto, L. et al. Single-molecule FRET studies on alpha-synuclein oligomerization of Parkinson’s disease genetically related mutants. Sci. Rep. 5, 16696 (2015).
Nath, S., Meuvis, J., Hendrix, J., Carl, S. A. & Engelborghs, Y. Early aggregation steps in α-synuclein as measured by FCS and FRET: evidence for a contagious conformational change. Biophys. J. 98, 1302–1311 (2010).
Degorce, F. et al. HTRF: a technology tailored for drug discovery — a review of theoretical aspects and recent applications. Curr. Chem. Genomics 3, 22–32 (2009).
Clarke, E. E. & Shearman, M. S. Quantitation of amyloid-β peptides in biological milieu using a novel homogeneous time-resolved fluorescence (HTRF) assay. J. Neurosci. Methods 102, 61–68 (2000).
Beaudet, L. et al. AlphaLISA immunoassays: the no-wash alternative to ELISAs for research and drug discovery. Nat. Methods 5, an8–an9 (2008).
Zhao, H. et al. AlphaLISA detection of alpha-synuclein in the cerebrospinal fluid and its potential application in Parkinson’s disease diagnosis. Protein Cell 8, 696–700 (2017).
Dale, N. C., Johnstone, E. K. M., White, C. W. & Pfleger, K. D. G. NanoBRET: the bright future of proximity-based assays. Front. Bioeng. Biotechnol. 7, 56 (2019).
Moriya, C. et al. PRDM14 directly interacts with heat shock proteins HSP90α and glucose-regulated protein 78. Cancer Sci. 109, 373–383 (2018).
van Ham, T. J. et al. Towards multiparametric fluorescent imaging of amyloid formation: studies of a YFP model of α-synuclein aggregation. J. Mol. Biol. 395, 627–642 (2010).
Chan, F. T. S., Kaminski, C. F. & Schierle, G. S. K. HomoFRET fluorescence anisotropy imaging as a tool to study molecular self-assembly in live cells. ChemPhysChem 12, 500–509 (2011).
Kundel, F. et al. Shedding light on aberrant interactions — a review of modern tools for studying protein aggregates. FEBS J. 285, 3604–3630 (2018).
De, S. & Klenerman, D. Imaging individual protein aggregates to follow aggregation and determine the role of aggregates in neurodegenerative disease. Biochim. Biophys. Acta Proteins Proteom. 1867, 870–878 (2019).
Horrocks, M. H. et al. Fast flow microfluidics and single-molecule fluorescence for the rapid characterization of α-synuclein oligomers. Anal. Chem. 87, 8818–8826 (2015).
Kjaergaard, M. et al. Oligomer diversity during the aggregation of the repeat region of tau. ACS Chem. Neurosci. 9, 3060–3071 (2018).
Anderson, V. L. & Webb, W. W. Transmission electron microscopy characterization of fluorescently labelled amyloid β 1-40 and α-synuclein aggregates. BMC Biotechnol. 11, 125 (2011).
Outeiro, T. F. et al. Formation of toxic oligomeric α-synuclein species in living cells. PLoS ONE 3, e1867 (2008).
Kiechle, M. et al. In vivo protein complementation demonstrates presynaptic α-synuclein oligomerization and age-dependent accumulation of 8–16-mer oligomer species. Cell Rep. 29, 2862–2874 (2019).
Frey, B., AlOkda, A. & Lashuel, H. Monitoring alpha-synuclein oligomerization and inclusion formation using bimolecular fluorescence complementation assays: what you see is not always what you get. J. Neurochem. 56, 583–586 (2020).
Mannini, B. et al. Stabilization and characterization of cytotoxic Aβ40 oligomers isolated from an aggregation reaction in the presence of zinc ions. ACS Chem. Neurosci. 9, 2959–2971 (2018).
Horrocks, M. H. et al. Single-molecule imaging of individual amyloid protein aggregates in human biofluids. ACS Chem. Neurosci. 7, 399–406 (2016).
Cliffe, R. et al. Filamentous aggregates are fragmented by the proteasome holoenzyme. Cell Rep. 26, 2140–2149 (2019).
Needham, L.-M. et al. ThX – a next-generation probe for the early detection of amyloid aggregates. Chem. Sci. 11, 4578–4583 (2020).
Bongiovanni, M. N. et al. Multi-dimensional super-resolution imaging enables surface hydrophobicity mapping. Nat. Commun. 7, 13544 (2016).
Lee, J.-E. et al. Mapping surface hydrophobicity of α-synuclein oligomers at the nanoscale. Nano Lett. 18, 7494–7501 (2018).
Kravchenko, K. et al. Analysis of anticoagulants for blood-based quantitation of amyloid β oligomers in the sFIDA assay. Biol. Chem. 398, 465–475 (2017).
Kühbach, K. et al. Application of an amyloid beta oligomer standard in the sFIDA assay. Front. Neurosci. 10, 8 (2016).
Kulawik, A., Heise, H., Zafiu, C., Willbold, D. & Bannach, O. Advancements of the sFIDA method for oligomer-based diagnostics of neurodegenerative diseases. FEBS Lett. 592, 516–534 (2018).
Hulsemann, M. et al. Biofunctionalized silica nanoparticles: standards in amyloid-β oligomer-based diagnosis of Alzheimer’s disease. J. Alzheimers Dis. 54, 79–88 (2016).
Herrmann, Y. et al. sFIDA automation yields sub-femtomolar limit of detection for Aβ aggregates in body fluids. Clin. Biochem. 50, 244–247 (2016).
Jain, A. et al. Probing cellular protein complexes using single-molecule pull-down. Nature 473, 484–488 (2011).
Aggarwal, V. & Ha, T. Single-molecule fluorescence microscopy of native macromolecular complexes. Curr. Opin. Struct. Biol. 41, 225–232 (2016).
Je, G. et al. Endogenous alpha-synuclein protein analysis from human brain tissues using single-molecule pull-down assay. Anal. Chem. 89, 13044–13048 (2017).
Mitkevich, O. V. et al. DNA aptamers detecting generic amyloid epitopes. Prion 6, 400–406 (2012).
Rahimi, F. Aptamers selected for recognizing amyloid β-protein — a case for cautious optimism. Int. J. Mol. Sci. 19, 668 (2018).
Schnitzbauer, J., Strauss, M. T., Schlichthaerle, T., Schueder, F. & Jungmann, R. Super-resolution microscopy with DNA-PAINT. Nat. Protoc. 12, 1198–1228 (2017).
Whiten, D. R. et al. Nanoscopic characterisation of individual endogenous protein aggregates in human neuronal cells. ChemBioChem 19, 2033–2038 (2018).
Kundel, F. et al. Hsp70 inhibits the nucleation and elongation of tau and sequesters tau aggregates with high affinity. ACS Chem. Biol. 13, 636–646 (2018).
Stroud, J. C., Liu, C., Teng, P. K. & Eisenberg, D. Toxic fibrillar oligomers of amyloid-β have cross-β structure. Proc. Natl Acad. Sci. USA 109, 7717–7722 (2012).
Ahmed, M. et al. Structural conversion of neurotoxic amyloid-β1–42 oligomers to fibrils. Nat. Struct. Mol. Biol. 17, 561–567 (2010).
Chen, S. W. et al. Structural characterization of toxic oligomers that are kinetically trapped during α-synuclein fibril formation. Proc. Natl Acad. Sci. USA 112, E1994–E2003 (2015).
Pieri, L., Madiona, K. & Melki, R. Structural and functional properties of prefibrillar α-synuclein oligomers. Sci. Rep. 6, 24526 (2016).
Karikari, T. K. et al. Preparation of stable tau oligomers for cellular and biochemical studies. Anal. Biochem. 566, 67–74 (2019).
Zhang, W. et al. Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer’s and Pick’s diseases. eLife 8, e43584 (2019).
Falcon, B. et al. Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature 561, 137–140 (2018).
Fitzpatrick, A. W. P. et al. Cryo-EM structures of tau filaments from Alzheimer’s disease brain. Nature 547, 185–190 (2017).
Guerrero-Ferreira, R. et al. Cryo-EM structure of alpha-synuclein fibrils. eLife 7, e36402 (2018).
Tian, Y., Liang, R., Kumar, A., Szwedziak, P. & Viles, J. H. 3D-visualization of amyloid-β oligomer and fibril interactions with lipid membranes by cryo-electron tomography. Preprint at bioRxiv https://doi.org/10.1101/2020.07.21.214072 (2020).
Cooper, M. A. Optical biosensors: where next and how soon? Drug Discov. Today 11, 1061–1067 (2006).
Aprile, F. A. et al. Selective targeting of primary and secondary nucleation pathways in Aβ42 aggregation using a rational antibody scanning method. Sci. Adv. 3, e1700488 (2017).
Chang, P.-T. et al. A newly designed molecule J2326 for Alzheimer’s disease disaggregates amyloid fibrils and induces neurite outgrowth. Neuropharmacology 92, 146–157 (2015).
Mustafa, M. K. et al. Detection of β-amyloid peptide (1–16) and amyloid precursor protein (APP770) using spectroscopic ellipsometry and QCM techniques: a step forward towards Alzheimers disease diagnostics. Biosens. Bioelectron. 26, 1332–1336 (2010).
Chauhan, N., Maekawa, T. & Kumar, D. N. S. Graphene based biosensors — accelerating medical diagnostics to new-dimensions. J. Mater. Res. 32, 2860–2882 (2017).
Jang, S. J., Lee, C. S. & Kim, T. H. α-Synuclein oligomer detection with aptamer switch on reduced graphene oxide electrode. Nanomaterials 10, 832 (2020).
Peña-Bahamonde, J., Nguyen, H. N., Fanourakis, S. K. & Rodrigues, D. F. Recent advances in graphene-based biosensor technology with applications in life sciences. J. Nanobiotechnol. 16, 75 (2018).
Arter, W. E., Levin, A., Krainer, G. & Knowles, T. P. J. Microfluidic approaches for the analysis of protein–protein interactions in solution. Biophys. Rev. 12, 575–585 (2020).
Yates, E. V. et al. Latent analysis of unmodified biomolecules and their complexes in solution with attomole detection sensitivity. Nat. Chem. 7, 802–809 (2015).
Arter, W. E. et al. Combining affinity selection and specific ion mobility for microchip protein sensing. Anal. Chem. 90, 10302–10310 (2018).
Arter, W. E. et al. Rapid fractionation and characterisation of alpha-synuclein oligomers in solution. Preprint at bioRxiv https://doi.org/10.1101/2020.03.10.985804 (2020).
Aebersold, R. & Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 537, 347–355 (2016).
Bader, J. M. et al. Proteome profiling in cerebrospinal fluid reveals novel biomarkers of Alzheimer’s disease. Mol. Syst. Biol. 16, e9356 (2020).
de Souza, N. & Picotti, P. Mass spectrometry analysis of the structural proteome. Curr. Opin. Struct. Biol. 60, 57–65 (2020).
Marx, V. A dream of single-cell proteomics. Nat. Methods 16, 809–812 (2019).
Bernstein, S. L. et al. Amyloid-β protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer’s disease. Nat. Chem. 1, 326–331 (2009). This article introduces the use of mass spectrometry to characterize oligomer populations.
Lieblein, T. et al. Structural rearrangement of amyloid-β upon inhibitor binding suppresses formation of Alzheimer’s disease related oligomers. eLife 9, e59306 (2020).
Österlund, N., Moons, R., Ilag, L. L., Sobott, F. & Gräslund, A. Native ion mobility-mass spectrometry reveals the formation of β-barrel shaped amyloid-β hexamers in a membrane-mimicking environment. J. Am. Chem. Soc. 141, 10440–10450 (2019).
Ilitchev, A. I. et al. Hetero-oligomeric amyloid assembly and mechanism: prion fragment PrP(106–126) catalyzes the islet amyloid polypeptide β-hairpin. J. Am. Chem. Soc. 140, 9685–9695 (2018).
Hoffmann, W. et al. NFGAIL amyloid oligomers: the onset of beta-sheet formation and the mechanism for fibril formation. J. Am. Chem. Soc. 140, 244–249 (2018).
Smith, A. M., Jahn, T. R., Ashcroft, A. E. & Radford, S. E. Direct observation of oligomeric species formed in the early stages of amyloid fibril formation using electrospray ionisation mass spectrometry. J. Mol. Biol. 364, 9–19 (2006).
Nakamura, A. et al. High performance plasma amyloid-β biomarkers for Alzheimer’s disease. Nature 554, 249–254 (2018).
Chang, E. & Kuret, J. Detection and quantification of tau aggregation using a membrane filter assay. Anal. Biochem. 373, 330–336 (2008).
Nasir, I., Linse, S. & Cabaleiro-Lago, C. Fluorescent filter-trap assay for amyloid fibril formation kinetics in complex solutions. ACS Chem. Neurosci. 6, 1436–1444 (2015).
Ruggeri, F. S., Habchi, J., Cerreta, A. & Dietler, G. AFM-based single molecule techniques: unraveling the amyloid pathogenic species. Curr. Pharm. Des. 22, 3950–3970 (2016).
Ruggeri, F. S., Šneideris, T., Vendruscolo, M. & Knowles, T. P. J. Atomic force microscopy for single molecule characterisation of protein aggregation. Arch. Biochem. Biophys. 664, 134–148 (2019).
Ruggeri, F. S. et al. Microfluidic deposition for resolving single-molecule protein architecture and heterogeneity. Nat. Commun. 9, 3890 (2018).
Qamar, S. et al. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation–π interactions. Cell 173, 720–734 (2018).
Ruggeri, F. S., Mannini, B., Schmid, R., Vendruscolo, M. & Knowles, T. P. J. Single molecule secondary structure determination of proteins through infrared absorption nanospectroscopy. Nat. Commun. 11, 2945 (2020). The combination of atomic force microscopy with infrared spectroscopy enables one to structurally characterize single molecules.
Yang, B., Liu, Z., Liu, H. & Nash, M. A. Next generation methods for single-molecule force spectroscopy on polyproteins and receptor–ligand complexes. Front. Mol. Biosci. 7, 85 (2020).
Sonn-Segev, A. et al. Quantifying the heterogeneity of macromolecular machines by mass photometry. Nat. Commun. 11, 1772 (2020).
Young, G. et al. Quantitative mass imaging of single biological macromolecules. Science 360, 423–427 (2018). This work introduces the use of interferometric scattering microscopy for single-molecule detection.
Young, G. & Kukura, P. Interferometric scattering microscopy. Annu. Rev. Phys. Chem. 19, 4827–4835 (2019).
Soltermann, F. et al. Quantifying protein–protein interactions by molecular counting with mass photometry. Angew. Chem. Int. Ed. 59, 10774–10779 (2020).
Wilham, J. M. et al. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog. 6, e1001217 (2010).
Atarashi, R. et al. Ultrasensitive human prion detection in cerebrospinal fluid by real-time quaking-induced conversion. Nat. Med. 17, 175–178 (2011).
Saborio, G. P., Permanne, B. & Soto, C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 411, 810–813 (2001).
Candelise, N. et al. Seeding variability of different alpha synuclein strains in synucleinopathies. Ann. Neurol. 85, 691–703 (2019).
De Luca, C. M. G. et al. Efficient RT-QuIC seeding activity for α-synuclein in olfactory mucosa samples of patients with Parkinson’s disease and multiple system atrophy. Transl Neurodegener. 8, 24 (2019).
van Rumund, A. et al. α-Synuclein real-time quaking-induced conversion in the cerebrospinal fluid of uncertain cases of parkinsonism. Ann. Neurol. 85, 777–781 (2019).
Garrido, A., Fairfoul, G., Tolosa, E. S., Martí, M. J. & Green, A. α-synuclein RT-QuIC in cerebrospinal fluid of LRRK2-linked Parkinson’s disease. Ann. Clin. Transl Neurol. 6, 1024–1032 (2019).
Metrick, M. A. et al. A single ultrasensitive assay for detection and discrimination of tau aggregates of Alzheimer and Pick diseases. Acta Neuropathol. Commun. 8, 22 (2020). This work illustrates the use of real-time quaking-induced conversion (RT-QuIC), a seed amplification assay, to discriminate different tauopathies.
Kraus, A. et al. Seeding selectivity and ultrasensitive detection of tau aggregate conformers of Alzheimer disease. Acta Neuropathol. 137, 585–598 (2019).
Shahnawaz, M. et al. Development of a biochemical diagnosis of Parkinson disease by detection of α-synuclein misfolded aggregates in cerebrospinal fluid. JAMA Neurol. 74, 163–172 (2017).
Bongianni, M. et al. α-Synuclein RT-QuIC assay in cerebrospinal fluid of patients with dementia with Lewy bodies. Ann. Clin. Transl Neurol. 6, 2120–2126 (2019).
Becker, K. et al. Detecting alpha synuclein seeding activity in formaldehyde-fixed MSA patient tissue by PMCA. Mol. Neurobiol. 55, 8728–8737 (2018).
Arosio, P., Cukalevski, R., Frohm, B., Knowles, T. P. J. & Linse, S. Quantification of the concentration of Aβ42 propagons during the lag phase by an amyloid chain reaction assay. J. Am. Chem. Soc. 236, 219–225 (2013).
Hooker, J. M. & Carson, R. E. Human positron emission tomography neuroimaging. Annu. Rev. Biomed. Eng. 21, 551–581 (2019).
Sevigny, J. et al. The antibody aducanumab reduces Aβ plaques in Alzheimer’s disease. Nature 537, 50–56 (2016).
Leuzy, A. et al. Tau PET imaging in neurodegenerative tauopathies — still a challenge. Mol. Psychiatry 24, 1112–1134 (2019).
Wong, D. F. et al. Characterization of 3 novel tau radiopharmaceuticals, 11C-RO-963, 11C-RO-643, and 18F-RO-948, in healthy controls and in Alzheimer subjects. J. Nucl. Med. 59, 1869–1876 (2018).
Betthauser, T. J. et al. In vivo characterization and quantification of neurofibrillary tau PET radioligand 18F-MK-6240 in humans from Alzheimer disease dementia to young controls. J. Nucl. Med. 60, 93–99 (2019).
Khoury, R. & Ghossoub, E. Diagnostic biomarkers of Alzheimer’s disease: a state-of-the-art review. Biomark. Neuropsychiatry 1, 100005 (2019).
Sanchez-Catasus, C. A. et al. FDG-PET for prediction of AD dementia in mild cognitive impairment. A review of the state of the art with particular emphasis on the comparison with other neuroimaging modalities (MRI and perfusion SPECT). Curr. Alzheimer Res. 14, 127–142 (2017).
Sehlin, D. et al. Engineered antibodies: new possibilities for brain PET? Eur. J. Nucl. Med. Mol. Imaging 46, 2848–2858 (2019).
Sehlin, D. et al. Antibody-based PET imaging of amyloid beta in mouse models of Alzheimer’s disease. Nat. Commun. 7, 10759 (2016).
Limbocker, R. et al. Trodusquemine enhances Aβ42 aggregation but suppresses its toxicity by displacing oligomers from cell membranes. Nat. Commun. 10, 225 (2019).
De, S. et al. Different soluble aggregates of Aβ42 can give rise to cellular toxicity through different mechanisms. Nat. Commun. 10, 1541 (2019).
Kumar, S. T., Donzelli, S., Chiki, A., Syed, M. M. K. & Lashuel, H. A. A simple, versatile and robust centrifugation-based filtration protocol for the isolation and quantification of α-synuclein monomers, oligomers and fibrils: Towards improving experimental reproducibility in α-synuclein research. J. Neurochem. 153, 103–119 (2020).
Lasagna-Reeves, C. A. et al. Identification of oligomers at early stages of tau aggregation in Alzheimer’s disease. FASEB J. 26, 1946–1959 (2012).
Castillo-Carranza, D. L. et al. Passive immunization with tau oligomer monoclonal antibody reverses tauopathy phenotypes without affecting hyperphosphorylated neurofibrillary tangles. J. Neurosci. 34, 4260–4272 (2014).
Kayed, R. et al. Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol. Neurodegener. 2, 18 (2007).
Ponsel, D., Neugebauer, J., Ladetzki-Baehs, K. & Tissot, K. High affinity, developability and functional size: The holy grail of combinatorial antibody library generation. Molecules 16, 3675–3700 (2011).
Winter, G., Griffiths, A. D., Hawkins, R. E. & Hoogenboom, H. R. Making antibodies by phage display technology. Annu. Rev. Immunol. 12, 433–455 (1994).
Bradbury, A. R. M., Sidhu, S., Dübel, S. & McCafferty, J. Beyond natural antibodies: the power of in vitro display technologies. Nat. Biotechnol. 29, 245–254 (2011).
Munke, A. et al. Phage display and kinetic selection of antibodies that specifically inhibit amyloid self-replication. Proc. Natl Acad. Sci. USA 114, 6444–6449 (2017).
Morgado, I. et al. Molecular basis of β-amyloid oligomer recognition with a conformational antibody fragment. Proc. Natl Acad. Sci. USA 109, 12503–12508 (2012).
Spencer, S., Bethea, D., Raju, T. S., Giles-Komar, J. & Feng, Y. Solubility evaluation of murine hybridoma antibodies. mAbs 4, 319–325 (2012).
Wu, S. J. et al. Structure-based engineering of a monoclonal antibody for improved solubility. Protein Eng. Des. Sel. 23, 643–651 (2010).
Jain, T. et al. Biophysical properties of the clinical-stage antibody landscape. Proc. Natl Acad. Sci. USA 114, 944–949 (2017).
Pepinsky, R. B. et al. Improving the solubility of anti-LINGO-1 monoclonal antibody Li33 by isotype switching and targeted mutagenesis. Protein Sci. 19, 954–966 (2010).
Burkovitz, A., Sela-Culang, I. & Ofran, Y. Large-scale analysis of somatic hypermutations in antibodies reveals which structural regions, positions and amino acids are modified to improve affinity. FEBS J. 281, 306–319 (2014).
Sormanni, P., Aprile, F. A. & Vendruscolo, M. Rational design of antibodies targeting specific epitopes within intrinsically disordered proteins. Proc. Natl Acad. Sci. USA 112, 9902–9907 (2015).
Sormanni, P., Aprile, F. A. & Vendruscolo, M. Third generation antibody discovery methods: in silico rational design. Chem. Soc. Rev. 47, 9137–9157 (2018).
Sormanni, P., Aprile, F. A. & Vendruscolo, M. The CamSol method of rational design of protein mutants with enhanced solubility. J. Mol. Biol. 427, 478–490 (2015).
Sormanni, P. & Vendruscolo, M. Protein solubility predictions using the CamSol method in the study of protein homeostasis. Cold Spring Harb. Perspect. Biol. 11, a033845 (2019).
Aprile, F. A. et al. Rational design of a conformation-specific antibody for the quantification of Aβ oligomers. Proc. Natl Acad. Sci. USA 117, 13509–13518 (2020).
Zeng, F. et al. Versatile near-infrared fluorescent probe for in vivo detection of Aβ oligomers. Bioorg. Med. Chem. 28, 115559 (2020).
Teoh, C. L. et al. Chemical fluorescent probe for detection of Aβ oligomers. J. Am. Chem. Soc. 137, 13503–13509 (2015).
Yang, J. et al. Highly specific detection of Aβ oligomers in early Alzheimer’s disease by a near-infrared fluorescent probe with a ‘V-shaped’ spatial conformation. Chem. Commun. 56, 583–586 (2020).
Lv, G. et al. A novel near-infrared fluorescent probe for detection of early-stage Aβ protofibrils in Alzheimer’s disease. Chem. Commun. 56, 1625–1628 (2020).
McIntyre, P. G. How many drops of CSF is enough? Postgrad. Med. J. 83, 158 (2007).
Lindquist, M. Neuroimaging results altered by varying analysis pipelines. Nature 582, 36–37 (2020).
Cummings, J. The role of biomarkers in Alzheimer’s disease drug development. Adv. Exp. Med. Biol. 1118, 29–61 (2019).
R&D Systems. Luminex assays & instruments. R&D Systems https://www.rndsystems.com/products/luminex-assays-and-high-performance-assays (2021).
Sartorius AG. BLI technology. Sartorius https://www.fortebio.com/applications/bli-technology (2020).
Arosio, P. et al. Microfluidic diffusion analysis of the sizes and interactions of proteins under native solution conditions. ACS Nano 10, 333–341 (2016).
Gough, K. C., Rees, H. C., Ives, S. E. & Maddison, B. C. Methods for differentiating prion types in food-producing animals. Biology 4, 785–813 (2015).
Rowe, C. C. et al. Standardized expression of 18F-NAV4694 and 11C-PiB β-amyloid PET results with the centiloid scale. J. Nucl. Med. 57, 1233–1237 (2016).
Acknowledgements
This work was supported by the UK Medical Research Council. P.S. is supported by a Royal Society University Research Fellowship (URF\R1\201461).
Author information
Authors and Affiliations
Contributions
All authors contributed to the preparation of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kulenkampff, K., Wolf Perez, AM., Sormanni, P. et al. Quantifying misfolded protein oligomers as drug targets and biomarkers in Alzheimer and Parkinson diseases. Nat Rev Chem 5, 277–294 (2021). https://doi.org/10.1038/s41570-021-00254-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41570-021-00254-9
This article is cited by
-
Misfolded protein oligomers: mechanisms of formation, cytotoxic effects, and pharmacological approaches against protein misfolding diseases
Molecular Neurodegeneration (2024)
-
Deciphering the effect of phytosterols on Alzheimer’s disease and Parkinson’s disease: the mediating role of lipid profiles
Alzheimer's Research & Therapy (2024)
-
Antibody production and characterization of the nucleoprotein of sever fever with thrombocytopenia syndrome virus (SFTSV) for effective diagnosis of SFTSV
Virology Journal (2023)
-
Patients with isolated REM-sleep behavior disorder have elevated levels of alpha-synuclein aggregates in stool
npj Parkinson's Disease (2023)
-
Soluble and insoluble protein aggregates, endoplasmic reticulum stress, and vascular dysfunction in Alzheimer’s disease and cardiovascular diseases
GeroScience (2023)