Abstract
Since its identification, Saccharomyces eubayanus has been recognized as the missing parent of the lager hybrid, S. pastorianus. This wild yeast has never been isolated from fermentation environments, thus representing an interesting candidate for evolutionary, ecological and genetic studies. However, it is imperative to develop additional molecular genetics tools to ease manipulation and thus facilitate future studies. With this in mind, we generated a collection of stable haploid strains representative of three main lineages described in S. eubayanus (PB-1, PB-2 and PB-3), by deleting the HO gene using CRISPR-Cas9 and tetrad micromanipulation. Phenotypic characterization under different conditions demonstrated that the haploid derivates were extremely similar to their parental strains. Genomic analysis in three strains highlighted a likely low frequency of off-targets, and sequencing of a single tetrad evidenced no structural variants in any of the haploid spores. Finally, we demonstrate the utilization of the haploid set by challenging the strains under mass-mating conditions. In this way, we found that S. eubayanus under liquid conditions has a preference to remain in a haploid state, unlike S. cerevisiae that mates rapidly. This haploid resource is a novel set of strains for future yeast molecular genetics studies.
Similar content being viewed by others
Introduction
Model organisms have been used for decades in biological and molecular studies, and are central players in the major breakthroughs in biology1. In eukaryotes, the yeast Saccharomyces cerevisiae is a popular model organism for research across many fields, ranging from classical genetics, molecular biology, biochemistry, and cellular physiology to population and comparative genomics2. The widespread utilization of S. cerevisiae is primarily because of its short cell cycle, small genome size, high recombination rates and self-fertilization capacity3. These features allowed researchers to generate different tools to simplify the genetic manipulation of yeast, such as in the large-scale development of null mutants4,5,6,7, and the generation of haploid and diploid strains with distinctive auxotrophic markers in different genetic backgrounds8,9,10,11. The generation of these collections was prompted by the easy manipulation of conventional genetic recombination approaches, and took advantage of the high homologous recombination (HR) rate in S. cerevisiae12,13. However, these approaches require the utilization of selective markers, drug-selectable markers and/or auxotrophic nutritional markers14,15. The use of auxotrophic markers can cause fitness changes, confounding the phenotypic effects of gene deletion15,16. Furthermore, there is a limited number of drug-selectable markers, restricting the simultaneous study of the fitness effects in null-mutants of multiple genes15. Although these selective markers can be recycled by recombinase-based methods such as Cre/LoxP or Flp/FRT, these techniques are time-consuming, since additional transformation rounds are required to remove selective markers14,17,18. In addition, these techniques have the drawback of leaving a copy of a repeat sequence in the genome, which can generate genome instability after multiple rounds of marker recovery14,19. An alternative to these laborious procedures is the type II bacterial Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) associated to the Cas9 protein (CRISPR-Cas9 system), allowing targeted double-strand breaks (DSBs) and simultaneous gene editing of all copies of the target sequence14,20,21. Since HR is the dominant mode of DSB repair in Saccharomyces yeast, short single-stranded- or double-stranded-DNA donor oligos can be used as a repair template, allowing targeted genome edition14. Different studies have precisely designed point mutations, single and multiple gene deletions, and multiplexed genome modifications at different loci, representing an ideal system for targeted mutagenesis20,22,23. In this sense, the CRISPR-Cas9 system has the advantage of both ease of use and broad applicability.
The S. cerevisiae collections have prompted different genetic experiments, such as the description of a gene’s function7, quantitative trait loci (QTLs) analysis and validation24,25,26,27, and gene expression analysis28,29. However, S. cerevisiae only represents a small fraction of the overall genetic and phenotypic diversity in the Saccharomyces genus30. Therefore, recent bioprospecting efforts have described large sets of strains in other Saccharomyces species, shifting the focus towards novel population genomics studies, including genome evolution, ecology and biotechnology. Moreover, novel Saccharomyces strains have a potential utilization in fermentation processes, using intra- or interspecies hybrids, and so these organisms are of interest for large-scale phenomics studies30,31,32,33. For example, during the past decade, Saccharomyces eubayanus and Saccharomyces jurei have been described as novel species in the Saccharomyces genus, where both species exhibit a great potential for brewing and dough fermentation34,35,36,37,38,39,40.
S. eubayanus has gained relevance since its isolation and identification in 2011 in the Argentinian Patagonia, as it is the missing parental species of the allopolyploid hybrid Saccharomyces pastorianus41. This hybrid is responsible for lager beer fermentation, and is currently used for the production of more than 90% of global commercial beer42. However, lager strains have a limited genetic diversity, which translates into a reduced variety of lager beers and low organoleptic differentiation31,43. Therefore, the isolation of S. eubayanus represented a promising step towards the generation of new brewing strains, either directly or through the generation of new Saccharomyces hybrids. Recently, several groups have reported the isolation of different S. eubayanus strains, including in China44, North America45, New Zealand46, and more recently, in Chile36. Phylogenetic analysis showed that this species has a wide genetic diversity, with five different lineages, where those from Patagonia have the most marked global genetic diversity compared to Holarctic and Chinese lineages36,37,47,48. Furthermore, this genetic diversity translates into an interesting phenotypic diversity, including phenotypes related to brewing conditions34,36. In this context, considering that S. eubayanus has been isolated from wild environments, and to date, there is no evidence of S. eubayanus isolation from fermentative environments, this yeast is thus an interesting candidate to become a valuable organism for evolutionary, ecological and genetic studies32. However, in order to adequately use S. eubayanus in molecular genetic studies, it is necessary to generate suitable genetic tools to facilitate its manipulation in the laboratory. Strains of S. eubayanus are diploid, highly homozygous and probably homothallic36,49. Thus, the generation of stable haploid versions representative of a broader genetic diversity in the species is an important step towards promoting the study of this biotechnologically relevant microorganism. In this sense, previous studies generated heterothallic Holarctic and admixed North American S. eubayanus strains for different biological studies50,51,52.
In this study, we developed a collection of 57 isogenic haploid strains representative of three Patagonian lineages described in the species. For this, we deleted the HO locus using the CRISPR-Cas9 system. The selected haploid strains from both mating types generated in this study were characterized under different conditions, observing that most spores mimic the diploid parental phenotype. Moreover, we sequenced a whole tetrad using nanopore sequencing to identify possible Cas9 off-targets and meiotic rearrangements. Finally, as proof of concept, we performed two experimental approaches for the generation of intraspecific hybrids under rich media, finding striking differences in mating frequency compared to S. cerevisiae. This new collection of S. eubayanus haploid strains is a valuable community resource that complements previously generated for molecular genetics studies.
Results and discussion
Generation of a large collection of haploid derivates in S. eubayanus
S. eubayanus is a diploid yeast, homothallic, and has been isolated mainly from wild environments36,49. To generate heterothallic strains, we decided to knock out the HO gene in wild diploid strains from different lineages (Fig. 1a). The HO gene encodes a site-specific endonuclease that promotes mating-type switching from a to α, or vice versa, in haploid strains, and its deletion has been widely used for the generation of stable haploid cells in yeast8,11,53. To delete the HO gene, we used a plasmid expressing the Cas9 enzyme and a gRNA cloning site developed by Fleiss et al.54; however, this plasmid had only been used in S. cerevisiae, without evaluating its efficiency in other Saccharomyces species.
Generation of a large set of stable haploid S. eubayanus strains. (a) Experimental procedure designed to generate the set of stable haploid S. eubayanus strains. A1. HO deletion, A2. Dissection of stable haploids and A3. Phenotyping of haploid strains. (b) Number of strains present in the wild collection of S. eubayanus, the sub-sets of null mutants and haploid strains generated (Created with BioRender.com). A single ‘MATa’ and ‘MATα’ spore was selected for phenotyping (c) Histogram of spore viability in 36 successfully transformed isolates (Δho).
We first evaluated the efficiency of the CRISPR-Cas9 system and the plasmid pAEF5 to obtain null mutants for the HO gene in the CL216.1 strain of S. eubayanus using four different gRNAs (gRNA1, gRNA2, gRNA3 and gRNA4, see methods), together with a 100-bp double stranded repair DNA fragment. HO gene deletion was observed in 11%, 0%, 42% and 0% of the investigated colonies for each gRNA (from 1 to 4), respectively. The deletion or mutation efficiencies obtained for S. eubayanus using CRISPR-Cas9 varies between studies, depending on the targeted region. In some cases, 100% efficiency was achieved in generating the targeted mutations (at the MALTX genes55), whilst in others, the efficiencies varied between 3 and 40%, depending on the gene and the genetic background56. Also, diploid yeast cells exhibit a barrier in utilizing DSB-mediated transformation schemes because diploid cells prefer homologous chromosomes for DSB repair, rather than exogenous linear DNA, resulting in a minority of recovered strains acquiring the targeted modification10. However, the efficiencies obtained in our study are in agreement with the efficiencies reported elsewhere in different S. cerevisiae strains utilizing the CRISPR-based gene deletion approach (1.3–100%)21,23,57. Considering the diploid nature of S. eubayanus, CRISPR-Cas9 based strategies allowed us to mutate both alleles in a single transformation event without the need to insert a selection cassette into the genome20. CRISPR-Cas9 has been used successfully in a few studies in S. eubayanus, generating null mutants55, integrating and overexpressing genes58, and introducing loss-of-function SNP mutations56. Therefore, based on our initial findings, gRNA3 was selected to repeat this procedure in a greater number of strains.
We subsequently used a collection of S. eubayanus strains spanning three of the six main lineages in the species, together with a subset of admixed strains, comprising about 55% of the genetic diversity found in this yeast species36,37. In this way, the initial collection of strains for HO knockout considered 89 strains (Fig. 1b), of which 83 strains were previously isolated from 10 different locations in Chile belonging to the PB-1, PB-2 and PB-3 populations; admixed clusters (ADM), five North American strains that grouped in the PB cluster, and the reference strain from Argentina were also used (Supplementary Table S1). Overall, 59 of the 89 S. eubayanus isolates were successfully transformed with the plasmid pAEF5 (Fig. 1b), including 20 strains from PB-1, 11 from PB-2, 19 from PB-3 and nine ADM strains (Supplementary Table S1). S. eubayanus has been genetically modified using electroporation and the lithium acetate procedure, however, historically only three different genetic backgrounds have been mainly used55,56,58. In this context, this study achieved genetic editing on a broader panel of genetically-distinct S. eubayanus strains.
To obtain stable haploid derivates, transformants were sporulated and dissected by micromanipulation (Fig. 1a). Only tetrads with four viable spores were considered for MAT locus genotyping. In this way, we obtained MATa and MATα haploid versions for 57 out of the 59 parental strains containing the plasmid pAEF5 (Fig. 1b, Supplementary Table S1). Strains CL1002.1 and CL611.1 did not sporulate and we were unable to obtain haploid derivates. Furthermore, in strains CL704.2 and CL836.1 we did not obtain four viable spores from the same ascus; however, as we identified MATa and MATα versions in both cases, both were included in the final haploid collection. Finally, we evaluated spore viability in 36 out of the 57 successfully transformed isolates, obtaining spore viabilities between 15 and 100% (Fig. 1c). In all, only one isolate exhibited spore viabilities lower than 50%, six isolates presented spore viabilities between 50 and 75%, and 30 isolates had spore viabilities over 75%. The high proportion of strains able to sporulate (~ 97%) agrees with previous data observed in S. eubayanus41 and other wild Saccharomyces strains, for which high sporulation and spore viability levels have been reported8,59,60.
Phenotypic profiling demonstrates similar phenotypes among diploid and haploid strains
For each strain we characterized the phenotypic profile of one MATa and MATα haploid derivates and their parental strains under eight different microculture conditions, including: 2% glucose at 25, 30 and 34 °C; 1.25 mM NaCl; 2% maltose; 5% and 10% ethanol; and 2% glycerol (Supplementary Table S2). In general, we observed that for most conditions and genetic backgrounds, there were no statistically significant differences in maximum growth rates between haploid strains and their respective parental strain (Fig. 2a). In this sense, we observed that in S. eubayanus most haploids and diploids exhibited similar performance under the environmental conditions tested in this study, where no general fitness advantages were evident in either ploidy state. Previously reported observations in S. cerevisiae and S. paradoxus using a more extensive set of conditions and strains also demonstrated no differences between either haploidy or diploidy61. However, we did find certain cases where one or both haploid versions showed statistically significant differences under certain conditions (Supplementary Table S2). We found specific ploidy x environment interactions, particularly for ethanol tolerance, where Mata haploid strains exhibited significantly lower growth rates than diploid strains (p-value < 0.05, Fig. 2b). The trend for the opposite mating type version MATα was similar, however, we found marginally significant differences in this case (p-value = 0.06, Student t-test). Indeed, DNA damage agents were previously shown to favor diploid asexual proliferation in S. cerevisiae and S. paradoxus compared to haploid strains61.
Phenotypic characterization of haploid strains and their respective diploid parental strains. (a) Maximum specific growth rates of diploids and haploids comparing all strains, ploidy, and environments. Line indicates the 1:1 correlation. (b) Maximum specific growth rates for diploids and haploids for all strains under 5% ethanol. Plotted values correspond to mean value of each strain. (*) depicts different levels of significance between haploid and diploid strains (Student t-test; *p ≤ 0.05 and ns: non-significant). (c) Principal component analysis (PCA) using the maximum specific growth rates under eight growth conditions, together with the distribution of individual haploid and diploid strains. Arrows depict the different environmental conditions (d) Hierarchically clustered heatmap of phenotypic diversity within haploid and diploid parental strains. Phenotypic values are calculated as normalized z-scores.
To analyze the whole phenotypic landscape in the set of 57 diploid and haploids strains, we performed a PCA analysis (Fig. 2c). The PC1 and PC2 components explain 33.8% and 15% of the observed variance, respectively, and combined, the two components account for 48.8% of the overall variation. PCA shows that growth in glycerol correlates negatively with the rest of the phenotypes where the carbon source was glucose or maltose. Interestingly, the individual factor map indicates no significant separation pattern according to ploidy and/or haploid strain mating-type, suggesting comparable phenotypes among diploid and haploid strains.
In addition, we also performed hierarchical clustering of the phenotypic data (Fig. 2d), observing three main clusters. Notably, 60% of the parental strains (34) clustered together with their two haploid versions, 18% (10) of the diploid parents clustered with one of the haploid versions, while 23% (13) distributed across different clusters compared to their two haploid versions. These results corroborate that most of our haploid strains are representative of their parental genetic backgrounds, as previously described61. For example, the CL715.1 strain and its haploid variants did not show statistically significant differences in seven of the eight media evaluated. However, the alpha version showed statistically significant differences when cultivated in 10% ethanol (Supplementary Table S2). Phenotypic differences between haploid and parental strains have also been observed in other yeast collections9,11. Therefore, this extensive collection of S. eubayanus represents a valuable resource for molecular genetics studies.
Similar brewing fermentative profiles in haploid and diploid strains
Considering the relevance that S. eubayanus has gained in the last decade for lager beer fermentation, we evaluated the fermentative profiles of parental strains and their haploid versions under conditions of beer wort fermentation. For this, we selected four strains representative of different lineages (CL1106.1 (ADM), CL216.1 (PB-3), CL601.1 (PB-3) and CL715.1 (PB-2)) for a 15-day lager fermentation in a 12°Brix wort and monitored the fermentation outcome by estimating CO2 loss. Overall, we did not observe statistically significant differences in the total CO2 output between haploid strains and their respective parental strains, except for the CL601.1-a strain (Fig. 3). All strains completely consumed the sugars present in the beer wort, except for maltotriose, which was previously reported as not being metabolized by S. eubayanus62. The haploid strains of CL216.1 and CL715.1-a showed significantly higher ethanol production than their respective parental strains, with differences of 0.74% (t-test, p < 0.001) and 0.64% (t-test, p < 0.001) for CL216.1-a and CL216.1-α strains, respectively, and 0.48% (t-test, p < 0.05) for the CL715.1-a strain (Supplementary Table S3). In general, the results of the fermentation assay demonstrate that this set of haploid strains is representative of the phenotypic fermentative performance of their diploid parents. Obtaining phenotypically representative haploid variants of the wide diversity of S. eubayanus strains is of relevance for genetic and biotechnological studies. In particular, these strains can be used at the fermentative level, where genetic analyses explaining differences between strains at the phenotypic level have not been undertaken. For example, strains belonging to the PB-3 subpopulation have a higher fermentative capacity than strains belonging to PB-2 or PB-136, reaffirming the existence of natural genetic variants in wild strains resulting in phenotypic diversity. In this way, this collection of haploid strains represents a valuable tool to be considered, for example, in the identification of QTLs.
Fermentation performance of haploid and diploid strains. Total CO2 loss for CL1106.1, CL216.1, CL601.1 and CL715.1 strains and their haploid strains. Plotted values correspond to three biological replicates. The (*) represents different levels of significance between the phenotype of haploid strains and their respective parental strain (t-test; *p ≤ 0.05 and ns: non-significant).
Low levels of off-target mutations in sequenced haploid strains
Previous studies have described off-target effects resulting from the use of a CRISPR-Cas9 system in genome editing63. In this sense, to evaluate off-target mutations, we sequenced the genome of three haploid strains, i.e., one spore of CL715.1, CL216.1, and CL601.1 (strains previously evaluated under fermentation conditions), using high-coverage whole-genome sequencing (WGS, Supplementary Table S4). After mapping the reads to the S. eubayanus reference genome (CBS12357)55, we performed variant calling and compared each haploid strain with their corresponding wild-type genotype that had been previously obtained using WGS36 to discover novel off-target mutations. We initially found 6, 12, and 16 putative mutations in CL601.1, CL216.1, and CL715.1, respectively (Supplementary Table S5a). After manual inspection of the alignments, we confirmed two single-nucleotide mutations, both occurring in the CL216.1 haploid strain in chromosomes 4 and 8. Interestingly, the mutation located in chromosome 4 was upstream of the HO gene, within the donor DNA recombination region, where the new allele coincides with the reference sequence used to generate the donor DNA. Furthermore, the mutation at chromosome 8, located in the coding sequence of the PRO2 gene, was also observed in the CL216.1 diploid sequencing using long reads35, suggesting that this mutation arose before the CRISPR-Cas9 procedure, and was not detected in the original CL216.1 WGS36. Off-target mutations correspond to DNA modifications at unintended sites, such as SNPs, deletions, insertions or inversions63, and depend on gRNA and experimental conditions64. In Saccharomyces yeast, these off-target mutations could arise from NHEJ of endonuclease-mediated DSBs, by HR between the 100 bp DNA donor or by homologous sequences at other genomic locations similar to the target sites57. Studies have evaluated the presence of off-target mutations after genome editing in Saccharomyces yeasts, in which they have been shown to be rare in haploid or homozygous strains56,57,65,66. Furthermore, off-target effects are considered unlikely in a small genome such as that of yeast21, coinciding with the results obtained in our study.
We also used haploid WGS to validate heterozygotes sites previously determined after WGS of diploid strains36. Strikingly, we found that out of the 807, 720 and 942 heterozygous sites called after WGS of the diploid strains CL216.1, CL601.1 and CL715.1, respectively, 98% of them were again identified as heterozygous sites in the haploid strains (Supplementary Tables S5b, S5b and S5b), suggesting that most heterozygote calls in diploids were false positives, and do not represent real heterozygotes sites, but rather polymorphic repeated regions or technical sequencing artifacts. These results suggest that the CRISPR-Cas9 technique used in our study likely generated a low number of off-targets in the manipulated strains. These results should be confirmed by sequencing a more significant number of haploid derivates..
Although we found few heterozygous sites validated after WGS of the three haploid strains (16–29) (Supplementary Tables S5b, S5b and S5b), meiotic events could also lead to the generation of structural variants and/or chromosome mis-segregation. Hence, we sequenced each of the four spores corresponding to one complete tetrad of the CL216.1 strain using nanopore long-read sequencing (Supplementary Table S6). Long-reads further confirmed the low number of heterozygous sites occurring in S. eubayanus, limited to only one site that segregated in a clear 2:2 proportion among spores (Supplementary Fig. S1). The draft genomes of each spore showed the absence of structural variants among spore genomes and demonstrated a flawless meiotic event (Supplementary Fig. S2). In general, meiosis provides a large source of genetic diversity by rearranging allelic combinations67, which is important to consider when haploid strain collections are generated. The main source of genetic diversity is meiotic crossovers (COs), that are reciprocal exchanges of chromosome arms between homologous chromosomes and are crucial for accurate homolog segregation at meiotic division I, and their absence leads to mis-segregation of homologs and aneuploid gametes68. Another source of genetic diversity during meiosis is the generation of punctual mutations. These normally occur in meiosis at higher frequencies than in vegetatively grown cells of S. cerevisiae, as a result of DSB repair in meiosis during the recombination process69. In this sense, we did not observe aneuploid or structural variants in any of the four spores derived from the CL216.1 strain, or de novo mutations, suggesting correct COs and homolog segregation during the meiotic process in CL216.1.
Intra-specific hybridization is common in S. cerevisiae, but not in S. eubayanus
Finally, we sought to evaluate the utility of our haploid collection by conducting, as proof of concept, two approaches to generate intra-specific hybrids (Fig. 4). First, we selected five strains that are representative of the different S. eubayanus lineages (CL449.1, CL715.1, CL216.1, CL601.1 and CL1106.1). The “Matα” and “Mata” versions of each strain were mixed in equal proportions and grown for 34 generations (four transfers) in a defined medium, and the proportion of haploid and diploid cells determined after each transfer (Fig. 4a). We evaluated the generation of diploid strains by colony isolation and identification of mating-type after each transfer. We observed a drastic decrease in MATα colonies in all three replicates. In replicates 2 and 3, we observed a predominance of MATa colonies, while a predominance of diploid colonies was observed in replicate 1 (Fig. 5a). Interestingly, MATa and MATα haploid strains did not exhibit growth rate differences under similar microculture conditions, and therefore the differences observed are specific under the dynamic competitive environment. A selective advantage for a certain trait is strongly dependent on environmental dynamics, where antagonism or competition can impact a strain’s success70. Previous studies demonstrated that sterile haploid mutants could exhibit a growth advantage over wild-type haploid cells71 suggesting that expression of mating pathway genes could differently impact the fitness of MATa and MATα cells.
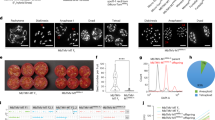
Generation of intraspecific hybrids in synthetic defined medium. (a) Proportion of diploid and haploid colonies identified during 34 generations. (b) SNPs identified in the eight diploid colonies genotyped for the DCR1 gene; colored squares represent the different parental strains and the hybrid’s genotype. (c) Mid-parent (left) and best-parent heterosis (right) in intraspecific hybrids evaluated under three different environmental conditions (2% glucose, 5% ethanol and 1.25 mM NaCl).
The diploid colonies in replicate 1 could originate from crosses between different strains, or crosses between two opposite mating types of the same genetic background. In addition, diploid colonies obtained from the same replicate may be clones from the same single hybridization event. To pinpoint the source of this variation, we genotyped the polymorphic DCR1 gene in eight diploid colonies to identify their respective parental strains (Fig. 5b and Supplementary Table S7). Five out of eight diploid colonies were identified as intraspecific hybrids: Three hybrids from replicate 1 (colony numbers 1, 3 and 4) and two from replicate 2 (colony numbers 6 and 7). Interestingly, in replicate 1 the CL449.1 strain predominated in both types of colonies, that is intraspecific hybrids and in self-crosses between strains of the same genetic background. On the other hand, CL715.1 strain predominated in replicates 2 and 3 in the two intraspecific hybrids (Supplementary Table S7).
To quantify the fitness of the hybrid-diploid colonies after the final serial transfer, we evaluated five, two and one diploid colonies from replicate 1, 2 and 3, respectively, under microculture conditions in 2% glucose, 5% ethanol and 1.25 mM NaCl, comparing their fitness against the five parental strains (Supplementary Table S8). We observed a significantly higher average growth rate (p-value < 0.05, ANOVA) in diploid colonies compared to parental strains in the three conditions evaluated, suggesting a greater fitness in these diploid individuals. Other diploid Saccharomyces intraspecific hybrids have shown superior phenotypes than their diploid parental strains, representing a successful strategy to survive under challenging conditions by creating phenotypic diversity72,73,74,75,76. Finally, we evaluated the heterosis coefficient in the five intraspecific hybrids relative to their respective parental strains in the same three conditions. In this case, we observed mild positive mid-parent and best-parent heterosis (heterosis coefficient > 1) (Fig. 5c). These results agree with previous studies, where intraspecific hybrids showed low levels of mid-parent heterosis for relative competitive growth, with no significant relationship between genetic divergence and heterosis77.
Considering the low intraspecific hybridization rate in S. eubayanus, we evaluated whether this situation depended on environmental conditions or the Saccharomyces species. For this, we carried out a second experimental approach for the generation of intraspecific hybrids by mass-mating under beer wort, as a representative alternative environmental condition. Furthermore, to determine whether the low hybridization rate was specific to S. eubayanus or reflected a more general trend in the Saccharomyces genus, we evaluated the hybridization rate in S. cerevisiae compared to S. eubayanus (Fig. 4b). We selected four strains representative of the PB-2, PB-3 and ADM lineages (CL715.1, CL216.1, CL601.1 and CL1106.1) in S. eubayanus, and four strains representative of the Wine/European, Sake, West African and North America lineages (DBVPG6765, Y12, DBVPG6044 and YPS128) in S. cerevisiae. The “MATα” and “MATa” versions of each strain were mixed and grown in malt extract for seven days and for 34 generations (four transfers). Subsequently, we determined the proportion of haploid and diploid cells after each transfer by colony PCR (Fig. 4b). As previously observed, in S. eubayanus, we identified a drastic decrease in MATα colonies, a predominance of MATa colonies, and the lack of hybrids after the fourth transfer in the three replicates (Fig. 6a).
On the other hand, S. cerevisiae quickly returns to a diploid state, which represent more than 50% of the colonies analyzed after the second transfer (Fig. 6b). These results suggest that the intraspecific hybridization rate depends on environmental conditions and the Saccharomyces species. The predominance of haploid colonies of S. eubayanus may be due to several factors. The presence of colonies of both mating-types after the first transfer, without the detection of diploid colonies, suggests that indeed the different strains of S. eubayanus are not mating, unlike in S. cerevisiae where after the first transfer, at least 30% of the colonies were diploid (Fig. 6b). Previous studies have demonstrated that cells of different mating types grown in a liquid medium can optimize their decision to mate or to proliferate by detecting the ratio of opposite-sex partners to same-sex competitors78. Furthermore, specific sexual aggregation via α/a-agglutinins is required for mating in a suspension of cells in liquid, which is determined by the probability of random mating encounters and the interaction strength of sexual α/a-agglutinins78,79. Indeed, the production of pheromones and the sexual aggregation capacity of S. eubayanus have not been studied; both factors can vary between strains, may be different from those reported for other Saccharomyces species, and may impact their mating capacity78,79. Finally, another factor to consider is the adaptive advantage of the ploidy level to different environments, which has been a matter of interest in studies of ecology and evolution80. Previous research has shown some adaptive advantages of haploid cells compared to their isogenic diploid strain under nutritional stress conditions61,81,82,83,84.
Conclusions
We obtained an extensive collection of haploid strains of S. eubayanus from different genetic backgrounds complementing previous resources generated for the species and the Saccharomyces genus. These strains were comprehensively phenotyped and represent a valuable and promising resource for studies in different aspects of biology, particularly for molecular genetic studies. Genomic analysis demonstrated the extremely low frequency of off-targets and no structural variants derived from the meiotic event. Finally, mass-mating experiments demonstrate the pertinence of the haploid collection for genetic studies and the preference of S. eubayanus to remain in the haploid state compared to S. cerevisiae, which quickly returns to a diploid state.
Methods
Strains
The S. eubayanus strains used in this study were selected from a collection of strains that have been published previously36,37. The strains comprised 83 Chilean strains collected from ten different localities36, one Argentinian strain41 and five North American strains45 (Supplementary Table S1). All strains were maintained on YPD solid medium (1% yeast extract, 2% peptone, 2% glucose, 2% agar). Yeast haploid strains are available upon request to the corresponding author.
Generation of null mutants by CRISPR-Cas9
HO null mutants were generated using a CRISPR-Cas9 method22 to generate stable haploid strains. The guide RNAs (gRNAs) were designed using the HO coding sequences from the reference strain CBS1235755 and the Benchling online tool (https://www.benchling.com/). We selected the four gRNAs with the highest “on-target” score, an optimized score for 20 bp gRNAs with NGG PAM, developed by Doench et al.64. The gRNAs were synthesized as single-stranded oligonucleotides (Macrogen, Korea), adding the SapI overhangs, 5’-ATC and 5’-AAC, and were phosphorylated (10 µM final concentration) using 10 units of T4 polynucleotide kinase (New England BioLabs, NEB) in a final volume of 10 µL at 37 °C for 1 h. Double-stranded oligonucleotides were generated by annealing equimolar amounts of phosphorylated single-stranded oligonucleotides under the following conditions: denaturation at 96 °C for 6 min and then cooling to 23 °C with a ramp of 0.1 °C/s. The four gRNAs were separately cloned in the plasmid pAEF554 (a gift from Gilles Fischer, Addgene plasmid #136,305), using standard “Golden Gate Assembly”85, digesting the plasmid with 10 units of SapI (NEB) and ligating this to each gRNA using 400 units of T4 DNA ligase (Promega) in a 10 µL final volume under the following conditions: 10 cycles at 42 °C for 2 min and 16 °C for 5 min, one cycle at 65 °C for 10 min and one cycle at 80 °C for 10 min. The product obtained was transformed into the E. coli DH5alpha competent (NEB, UK) following the manufacturer’s recommendations. The plasmids were recovered using a FavorPrep Plasmid DNA Extraction Mini Kit (Favorgen Biotech Corp.), quantified using a Qubit system (Thermo Fisher, Waltham, Massachusetts, US.) and verified in a 1.5% agarose gel.
The donor DNA consisted of a 100 bp synthetic double-stranded DNA fragment (Macrogen, Korea) containing flanking sequences to the target HO gene, with a sequence homology of 50 bp upstream of the start codon and 50 bp downstream of the stop codon. This donor DNA was amplified by PCR using 1 × Phusion High-Fidelity PCR Master Mix (Thermo Fisher), 0.52 µM of each primer and 4 µM of donor DNA under the following conditions: 98 °C for 30 s; 35 cycles at 98 °C for 10 s, 55 °C for 30 s and 72 °C for 20 s; 72 °C for 15 min.
Finally, each diploid strain was co-transformed with the plasmid carrying the gRNA together with the Cas9 gene, and the donor DNA using the standard lithium acetate protocol86 with temperature modifications. Briefly, the strains were grown overnight in 5 mL YPD under constant agitation at 20 °C. Then, 300 µL overnight culture were inoculated in 5 mL YPD and incubated under the same conditions for 4 h or until the culture reached 108 cells/mL. From this, 1 mL was washed three times with distilled water, then washed twice in 1 mL of lithium acetate 0.1 M (LiAc) and cells were recovered by centrifugation. The pellet of cells was mixed with 240 µL polyethylene glycol 3350 50% w/v (PEG 3350), 36 µL 1 M LiAc, 20 µL denatured salmon sperm DNA (10 mg/mL), 500 ng of a plasmid carrying the gRNA and the Cas9, and 24 µL of donor DNA for the repair of CRISPR-induced DSBs. The cells were resuspended by vortex mixing and incubated for 45 min at 20 °C. Then, a heat shock was performed at 37 °C for 20 min. After transformation, cells were washed with 1 mL distilled water and plated on YPD with 0.2 mg/mL hygromycin for selection. Plates were incubated between 3 to 5 days at 20 °C. Correct gene deletion was confirmed by colony PCR using GoTaq DNA polymerase (Promega). All primer, gRNAs and donor DNA sequences are listed in Supplementary Table S9.
Generation of stable haploid strains
Isogenic haploid strains were constructed by deleting the HO gene using a CRISPR-Cas9 method as described above. Initially, the four gRNAs were evaluated separately in the CL216.1 strain, determining the efficiency of each gRNA as the number of colonies with the proper deletion divided by the total number of colonies evaluated by colony PCR. The most efficient gRNA was used to transform the other strains in the collection. Diploid Δho cells were sporulated on 2% potassium acetate agar plates (2% agar) for at least seven days at 20 °C. Meiotic segregants were obtained by dissecting tetrad ascospores treated with 10 µL Zymolyase 100 T (50 mg/mL) on YPD agar plates with a SporePlay micromanipulator (Singer Instruments, UK). Spores from four viable spore tetrads were selected to determine the mating type by colony PCR of the MAT locus8 using GoTaq DNA Polymerase (Promega). All the primers used in these experimental procedures are listed in Supplementary Table S9.
Spore viability
Strains were sporulated on 2% potassium acetate agar plates as described above. Tetrad ascospores were treated with 10 µL Zymolyase 100 T (50 mg/mL) and dissected with a SporePlay micromanipulator. At least 20 ascospores per strain were dissected on YPD plates and incubated at 20 °C for four days. Spore viability was calculated using the formula (n° of viable spores/n° of dissected spores) × 100.
Genotypic characterization
Genomic DNA from a single spore of CL216.1-a, CL715.1-a and CL601.1-a was prepared for whole-genome sequencing using the Qiagen Genomic-tip 20/G kit (Qiagen, Germany) as previously described36 and sent for DNBseq sequencing (BGISEQ-G400 platform). Reads were filtered and trimmed using Fastp 0.20.1 (–3-l 50-cut_mean_quality 30)87. Cleaned reads were mapped to the S. eubayanus reference strain CBS12357 using BWA mem 0.7.1788, after which PCR duplicates were marked using Picard tools. Mapping files corresponding to the original diploid strains were obtained from36. Variants were jointly called on haploid and diploid alignments using freeBayes 1.3.189. Variants were analyzed in R and visually inspected using the IGV genome browser90.
To perform nanopore sequencing of a full tetrad of the CL216.1 strain, DNA was extracted from each spore using the Quick-DNA HMW MagBead Kit (Zymo Research). High molecular weight genomic DNA was purified using magnetic beads following the kit manufacturer’s protocol. DNA integrity was verified using an agarose gel and DNA quantity was assessed using Quantus (Promega, Madison, Wisconsin, US.). Libraries were prepared following the manufacturer's protocol, barcodes were added to each library, and these were sequenced on a single R9.4 flow cell using a Minion (Oxford Nanopore Technologies, UK). The raw fast5 files were transformed to fastq files and de-barcoded using Guppy v5.0.14 with the “super high accuracy” model. Long reads were mapped to the S. eubayanus reference CBS12357 with minimap2 (ax map-ont –secondary = no)91 and variants were called using PEPPER (–ont_r9_guppy5_sup)92. Long-read assemblies of each spore were generated using Flye v2.9 (–nano-hq –scaffold)93. To evaluate synteny, each assembly was compared using nucmer94. Structural variations were identified using MUM&Co95.
Phenotypic characterization
One MATa and MATα haploid version of each haploid strain and their respective diploid parental background were phenotyped under microculture conditions. For this, we estimated mitotic growth in 96-well plates under different conditions in three biological replicates, including: Yeast Nitrogen Base (YNB) supplemented with 2% glucose at 25 °C, 30 °C and 34 °C; 2% glycerol, 2% glucose and 1.25 mM NaCl; 2% maltose; 2% glucose and 5% ethanol; 2% glucose and 10% ethanol. All conditions were evaluated at 25 °C in 0.67% YNB medium (Difco, France). First, haploid and diploid strains were pre-grown in 200 µL YNB medium supplemented with 2% glucose for 48 h at 25 °C and used to inoculate a 96-well plate with a final volume of 200 µL at an initial OD600nm of 0.1. The growth curves were monitored by measuring the OD600nm every 30 min in a Tecan Sunrise absorbance microplate reader (Tecan Group Ltd., Switzerland). All experiments were carried out in triplicates. The maximum specific growth rate (µmax) was determined using GrowthRates software with default parameters96.
Fermentations were carried out in three biological replicates using previously oxygenated (15 mg/L) 12°Plato (°P) beer wort, supplemented with 0.3 ppm ZnCl2. The pre-cultures were grown in 5 mL 6°P wort for 24 h at 20 °C in constant agitation at 150 rpm. Inocula were then transferred to 50 mL 12°P wort and incubated for 24 h at 20 °C in constant agitation at 150 rpm. The cells were collected by centrifugation and used to calculate the final cell concentration for use in each fermentation according to the formula described by White and Zainasheff97. Cells were inoculated into 50 mL 12°P wort in 250 mL bottles and airlocks with 30% glycerol. The fermentations were incubated at 12 °C, with no agitation for 15 days and monitored by weighing the bottles daily to determine weight loss over time.
Metabolites quantification by HPLC
Glucose, fructose, maltose, maltotriose and ethanol concentrations were determined by High-Performance Liquid Chromatography (HPLC) after 15 days of fermentation as previously described35,48. Samples were obtained extracting 0.5 mL fermented beer wort and filtered using 0.22 μm filters. Filtered samples (20 µL) were injected in a Shimadzu Prominence HPLC (Shimadzu, USA) with a BioRad HPX-87H column using 5 mM sulfuric acid and 4 mL acetonitrile per liter of sulfuric acid as the mobile phase at a 0.5 mL/min flow rate98. Glucose, fructose, maltose and maltotriose uptake were estimated as the difference between the initial and final concentration before and after fermentation, respectively.
Generation of intra-specific hybrids by mass-mating
Five different genetic backgrounds were selected to generate intra-specific hybrids by mass-mating (CL216.1, CL601.1, CL715.1, CL1106.1 and CL449.1) (Fig. 4a). First, one colony from each S. eubayanus strain and mating type was cultured in 0.67% YNB medium (Difco, France) with 2% glucose at 25 °C in constant agitation at 150 rpm. Each pre-inoculum was then utilized to prepare a co-culture at an initial OD600nm of 0.1 of each strain in 150 mL 0.67% YNB medium with 2% glucose. This co-culture was divided in three 50 mL replicates and incubated at 25 °C in constant agitation at 150 rpm during 72 h. Subsequently, the cultures were used to inoculate fresh 50 mL cultures at an initial OD600nm of 0.1, and this procedure was sequentially repeated four times. After each incubation, colonies were isolated for each replicate on YPD solid medium and the mating type determined by colony PCR as described above (at least 10 colonies per replicate). The number of generations was determined using the formula log2(OD600 final/0.1)99.
Diploid colonies identified after the last incubation were characterized under microculture conditions as described above, together with the parental diploid and haploid strains. The conditions evaluated were: 2% glucose at 25 °C, 2% glucose with 1.25 mM NaCl, and 2% glucose with 5% ethanol. Mid-parent and best-parent heterosis were determined as previously described77,100, using Eqs. (1) and (2), where mid-parent heterosis denotes the hybrid deviation from the mid-parent performance and best-parent heterosis denotes the hybrid deviation from the better parent phenotypic value101.
where:
\({\mu max}_{h}\): growth rate of a hybrid.
Intra-specific hybrids by mass-mating were also generated under fermentation conditions (Fig. 4b). Four different genetic background were selected: CL216.1, CL601.1, CL715.1 and CL1106.1. The pre-culture conditions were the same as described above; each haploid strain was grown in 5 mL 6°P wort for 24 h at 20 °C in constant agitation at 150 rpm, then transferred to 50 mL 12°P wort and incubated for 24 h at 20 °C in constant agitation at 150 rpm. Each pre-inoculum was used to prepare a co-culture at a final concentration of 1 × 106 cells/mL and transferred to three replicates to obtain a final concentration of 1.5 × 106 cells/mL in 50 mL 12°P wort in 250 mL bottles and airlocks with 30% glycerol. The fermentations were incubated at 20 °C, with no agitation for 7 days and monitored by weighing the bottles daily to determine weight loss over time. At the end of fermentation, the cultures were used to inoculate fresh 50 mL 12°P wort at an inoculum density of 1.5 × 106 cells/mL, repeating the fermentation and re-inoculation four times. Also, we used four different S. cerevisiae strains that represent the main lineages described in the species: YPS128 (North American, NA), DBVPG6044 (West African, WA), Y12 (Sake, SA) and DBVPG6765 (Wine/European, WE) (Supplementary Table S10)8. The number of generations was determined using the formula log2(final cells/1.5 × 106)102. Finally, after each fermentation, the proportions of diploid and haploid cells were determined by colony PCR of colonies isolated for each replicate on solid YPD medium, as described above.
Hybrid genotyping
Diploid colonies identified after the last incubation in the mass-mating experiment were genotyped to determined their intraspecific hybrid status by sequencing the DCR1 gene45. Genomic DNA of eight diploid colonies was extracted using the Wizard® Genomic DNA Purification Kit (Promega) according to the manufacturer’s instructions. PCR reactions were performed in a final volume of 25 µL containing 1 × GoTaq DNA polymerase Master Mix (Promega), 0.2 µM of each primer and 1 µL DNA (25 ng/µL) under the following conditions: 95 °C for 2 min; 35 cycles at 95 °C for 15 s, 52–55 °C for 15 s and 72 °C for 90 s; 72 °C for 5 min. Sanger sequencing was performed in an Applied Biosystem 3130XL instrument (Pontificia Universidad Católica de Chile, Santiago, Chile). Sequences of diploid colonies and wild strains were aligned using a Multiple Sequence Comparison by Log-Expectation (MUSCLE) algorithm with default parameters in MEGA-X version 10.2.4. Sequences of wild strains used in the mass-mating experiment were previously published36.
Data and statistical analysis
Data visualization and statistical analyses were performed with R software version 4.0.3. The maximum specific growth rates and total CO2 loss were compared using an analysis of variance (ANOVA) and the mean values of the three replicates were statistically analyzed using Student’s t-test and corrected for multiple comparisons using the Benjamini–Hochberg method. A p-value less than 0.05 (p < 0.05) was considered statistically significant. A principal component analysis (PCA) was performed using the FactoMineR package version 2.4 to compute principal component methods and the factoextra package version 1.07 for extracting, visualizing and interpreting the results. Heatmaps was generated using the ComplexHeatmap package version 2.6.2.
Data availability
All fastq sequences were deposited in the National Center for Biotechnology Information (NCBI) as a Sequence Read Archive under the BioProject accession number PRJNA800259 (www.ncbi.nlm.nih.gov/bioproject/800259).
References
Alliance, T. & Consortium, G. R. The alliance of genome resources: Building a modern data ecosystem for model organism databases. Genetics 213, 1189–1196 (2019).
Cubillos, F. A. Exploiting budding yeast natural variation for industrial processes. Curr. Genet. 62, 745–751 (2016).
Pretorius, I. S. & Boeke, J. D. Yeast 2.0-connecting the dots in the construction of the world’s first functional synthetic eukaryotic genome. FEMS Yeast Res. 18, 1–15 (2018).
Replogle, K., Hovland, L. & Rivier, D. H. Designer deletion and prototrophic strains derived from Saccharomyces cerevisiae strain W303–1a. Yeast 15, 1141–1149 (1999).
Carter, Z. & Delneri, D. New generation of loxP-mutated deletion cassettes for the genetic manipulation of yeast natural isolates. Yeast 27, 765–775 (2010).
Akada, R. et al. PCR-mediated seamless gene deletion and marker recycling in Saccharomyces cerevisiae. Yeast 23, 399–405 (2006).
Giaever, G. & Nislow, C. The yeast deletion collection: A decade of functional genomics. Genetics 197, 451–465 (2014).
Cubillos, F. A., Louis, E. J. & Liti, G. Generation of a large set of genetically tractable haploid and diploid Saccharomyces strains. FEMS Yeast Res. 9, 1217–1225 (2009).
Louvel, H., Gillet-Markowska, A., Liti, G. & Fischer, G. A set of genetically diverged Saccharomyces cerevisiae strains with markerless deletions of multiple auxotrophic genes. Yeast 31, 91–101 (2014).
Alexander, W. G., Doering, D. T. & Hittinger, C. T. High-efficiency genome editing and allele replacement in prototrophic and wild strains of saccharomyces. Genetics 198, 859–866 (2014).
Coi, A. L., Legras, J. L., Zara, G., Dequin, S. & Budroni, M. A set of haploid strains available for genetic studies of Saccharomyces cerevisiae flor yeasts. FEMS Yeast Res. 16, 1–9 (2016).
Shao, S. et al. Enhancing CRISPR/Cas9-mediated homology-directed repair in mammalian cells by expressing Saccharomyces cerevisiae Rad52. Int. J. Biochem. Cell Biol. 92, 43–52 (2017).
McIlwraith, M. J. & West, S. C. DNA repair synthesis facilitates RAD52-mediated second-end capture during DSB repair. Mol. Cell 29, 510–516 (2008).
Rainha, J., Rodrigues, J. L. & Rodrigues, L. R. CRISPR-Cas9: A powerful tool to efficiently engineer saccharomyces cerevisiae. Life 11, 1–16 (2021).
Fraczek, M. G., Naseeb, S. & Delneri, D. History of genome editing in yeast. Yeast 35, 361–368 (2018).
Baganz, F., Hayes, A., Marren, D., Gardner, D. C. & Oliver, S. G. Suitability of replacement markers for functional analysis studies in Saccharomyces cerevisiae. Yeast 13, 1563–1573 (1997).
Park, Y., Masison, D., Eisenberg, E. & Greene, L. E. Application of the FLP/FRT system for conditional gene deletion in yeast Saccharomyces cerevisiae. Yeast 28, 673–681 (2011).
Yamanishi, M. & Matsuyama, T. A modified cre-lox genetic switch to dynamically control metabolic flow in Saccharomyces cerevisiae. ACS Synth. Biol. 1, 172–180 (2012).
Solis-Escalante, D. et al. Efficient simultaneous excision of multiple selectable marker cassettes using I-SceI-induced double-strand DNA breaks in Saccharomyces cerevisiae. FEMS Yeast Res. 14, 741–754 (2014).
Mans, R. et al. CRISPR/Cas9: A molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res. 15, 1–15 (2015).
Stovicek, V., Holkenbrink, C. & Borodina, I. CRISPR/Cas system for yeast genome engineering: Advances and applications. FEMS Yeast Res. 17, 1–16 (2017).
Dicarlo, J. E. et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucl. Acids Res. 41, 4336–4343 (2013).
Vigentini, I., Gebbia, M., Belotti, A., Foschino, R. & Roth, F. P. CRISPR/Cas9 system as a valuable genome editing tool for wine yeasts with application to decrease urea production. Front. Microbiol. 8, 1–11 (2017).
Jara, M. et al. Mapping genetic variants underlying differences in the central nitrogen metabolism in fermenter yeasts. PLoS ONE 9, e86533 (2014).
Cubillos, F. A. et al. High-resolution mapping of complex traits with a four-parent advanced intercross yeast population. Genetics 195, 1141–1155 (2013).
Salinas, F. et al. The genetic basis of natural variation in oenological traits in saccharomyces cerevisiae. PLoS ONE 7, e49640 (2012).
Cubillos, F. et al. Identification of nitrogen consumption genetic variants in yeast through QTL mapping and bulk segregant RNA-seq analyses. G3 Genes Genom. Gen. 7, 1693–1705 (2017).
Salinas, F. et al. Natural variation in non-coding regions underlying phenotypic diversity in budding yeast. Sci. Rep. 6, 21849 (2016).
Villarroel, C. A., Bastías, M., Canessa, P. & Cubillos, F. A. Uncovering divergence in gene expression regulation in the adaptation of yeast to nitrogen scarcity. mSystems 6, (2021).
Alsammar, H. & Delneri, D. An update on the diversity, ecology and biogeography of the Saccharomyces genus. FEMS Yeast Res. 20, (2020).
Cubillos, F. A., Gibson, B., Grijalva-Vallejos, N., Krogerus, K. & Nikulin, J. Bioprospecting for brewers: Exploiting natural diversity for naturally diverse beers. Yeast https://doi.org/10.1002/yea.3380 (2019).
Libkind, D. et al. Into the wild: new yeast genomes from natural environments and new tools for their analysis. FEMS Yeast Res. 20, (2020).
Molinet, J. & Cubillos, F. A. Wild yeast for the future: Exploring the use of wild strains for wine and beer fermentation. Front. Genet. 11, 1–8 (2020).
Urbina, K. et al. Volatile compound screening using HS-SPME-GC/MS on saccharomyces eubayanus strains under low-temperature pilsner wort fermentation. Microorganisms 8, 1–19 (2020).
Mardones, W. et al. Molecular profiling of beer wort fermentation diversity across natural Saccharomyces eubayanus isolates. Microb. Biotechnol. https://doi.org/10.1111/1751-7915.13545 (2020).
Nespolo, R. F. et al. An Out-of-Patagonia migration explains the worldwide diversity and distribution of Saccharomyces eubayanus lineages. PLoS Genet. 16, e1008777 (2020).
Langdon, Q. K. et al. Postglacial migration shaped the genomic diversity and global distribution of the wild ancestor of lager-brewing hybrids. PLOS Genet. 16, e1008680 (2020).
Giannakou, K. et al. Biotechnological exploitation of Saccharomyces jurei and its hybrids in craft beer fermentation uncovers new aroma combinations. Food Microbiol. 100, 103838 (2021).
Hutzler, M. et al. Unique brewing-relevant properties of a strain of saccharomyces jurei isolated from ash (Fraxinus excelsior). Front. Microbiol. 12, 1–14 (2021).
Magalhães, F., Calton, A., Heiniö, R. L. & Gibson, B. Frozen-dough baking potential of psychrotolerant Saccharomyces species and derived hybrids. Food Microbiol. 94, (2021).
Libkind, D. et al. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc. Natl. Acad. Sci. 108, 14539–14544 (2011).
Lin, C. L. et al. Packing a punch: understanding how flavours are produced in lager fermentations. FEMS Yeast Res. 21, 1–14 (2021).
Bonatto, D. The diversity of commercially available ale and lager yeast strains and the impact of brewer ’ s preferential yeast choice on the fermentative beer profiles. Food Res. Int. 141, 110125 (2021).
Bing, J., Han, P. J., Liu, W. Q., Wang, Q. M. & Bai, F. Y. Evidence for a far east asian origin of lager beer yeast. Curr. Biol. 24, R380–R381 (2014).
Peris, D. et al. Population structure and reticulate evolution of Saccharomyces eubayanus and its lager-brewing hybrids. Mol. Ecol. 23, 2031–2045 (2014).
Gayevskiy, V. & Goddard, M. R. Saccharomyces eubayanus and Saccharomyces arboricola reside in North Island native New Zealand forests. Environ. Microbiol. 18, 1137–1147 (2016).
Eizaguirre, J. I. et al. Phylogeography of the wild Lager-brewing ancestor (Saccharomyces eubayanus) in Patagonia. Environ. Microbiol. 20, 3732–3743 (2018).
Villarreal, P. et al. Identification of new ethanol-tolerant yeast strains with fermentation potential from central Patagonia. Yeast https://doi.org/10.1002/yea.3662 (2021).
Gibson, B. et al. New yeasts-new brews: Modern approaches to brewing yeast design and development. FEMS Yeast Res. 17, 1–13 (2017).
Baker, E. C. P. et al. Mitochondrial DNA and temperature tolerance in lager yeasts. Sci. Adv. 5, 1–8 (2019).
Brouwers, N. et al. Himalayan saccharomyces eubayanus genome sequences reveal genetic markers explaining heterotic maltotriose consumption by saccharomyces pastorianus hybrids. Appl. Environ. Microbiol. 85, 1–22 (2019).
Baker, E. C. P. & Hittinger, C. T. Evolution of a novel chimeric maltotriose transporter in Saccharomyces eubayanus from parent proteins unable to perform this function. PLoS Genet. 15, e1007786 (2019).
Karademir Andersson, A., Oredsson, S. & Cohn, M. Development of stable haploid strains and molecular genetic tools for Naumovozyma castellii (Saccharomyces castellii). Yeast 33, 633–646 (2016).
Fleiss, A. et al. Reshuffling yeast chromosomes with CRISPR/Cas9. PLOS Genet. 15, e1008332 (2019).
Brickwedde, A. et al. Structural, physiological and regulatory analysis of maltose transporter genes in Saccharomyces eubayanus CBS 12357T. Front. Microbiol. 9, 1–18 (2018).
Mertens, S. et al. Reducing phenolic off-flavors through CRISPR-based gene editing of the FDC1 gene in Saccharomyces cerevisiae x Saccharomyces eubayanus hybrid lager beer yeasts. PLoS ONE 14, e0209124 (2019).
Jakočinas, T. et al. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab. Eng. 28, 213–222 (2015).
Brouwers, N. et al. In vivo recombination of Saccharomyces eubayanus maltose-transporter genes yields a chimeric transporter that enables maltotriose fermentation. PLoS Genet. 15, 1–30 (2019).
Gallone, B. et al. Origins, evolution, domestication and diversity of Saccharomyces beer yeasts. Curr. Opin. Biotechnol. 49, 148–155 (2018).
Gerke, J., Lorenz, K. & Cohen, B. Genetic interactions between transcription factors cause natural variation in yeast. Science (80-) 323, 498–501 (2009).
Zörgö, E. et al. Ancient evolutionary trade-offs between yeast ploidy states. PLoS Genet. 9, (2013).
Hebly, M. et al. S. cerevisiae × S. eubayanus interspecific hybrid, the best of both worlds and beyond. FEMS Yeast Res. 15, 1–14 (2015).
Blattner, G., Cavazza, A., Thrasher, A. J. & Turchiano, G. Gene editing and genotoxicity: Targeting the off-targets. Front. Genome Ed. 2, 1–10 (2020).
Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Chin, Y. W., Shin, S. C., Han, S., Jang, H. W. & Kim, H. J. CRISPR/Cas9-mediated Inactivation of arginase in a yeast strain isolated from Nuruk and its impact on the whole genome. J. Biotechnol. 341, 163–167 (2021).
Gorter De Vries, A. R. et al. Allele-specific genome editing using CRISPR-Cas9 is associated with loss of heterozygosity in diploid yeast. Nucl. Acids Res. 47, 1362–1372 (2019).
Brion, C. et al. Variation of the meiotic recombination landscape and properties over a broad evolutionary distance in yeasts. PLoS Genet. 13, 1–21 (2017).
Martini, E. et al. Genome-wide analysis of heteroduplex DNA in mismatch repair-deficient yeast cells reveals novel properties of meiotic recombination pathways. PLoS Genet. 7, (2011).
Simchen, G., Mansour, O., Morciano, L., Zenvirth, D. & Arbel-Eden, A. Mutagenicity in haploid yeast meiosis resulting from repair of DSBs by the sister chromatid. Curr. Genet. 67, 799–806 (2021).
Onetto, C. A., Borneman, A. R. & Schmidt, S. A. Strain-specific responses by saccharomyces cerevisiae to competition by non-saccharomyces yeasts. Fermentation 7, (2021).
Lang, G. I., Murray, A. W. & Botstein, D. The cost of gene expression underlies a fitness trade-off in yeast. Proc. Natl. Acad. Sci. U. S. A. 106, 5755–5760 (2009).
Steensels, J., Meersman, E., Snoek, T., Saels, V. & Verstrepen, K. J. Large-scale selection and breeding to generate industrial yeasts with superior aroma production. Appl. Environ. Microbiol. 80, 6965–6975 (2014).
Stelkens, R. B., Brockhurst, M. A., Hurst, G. D. D. & Greig, D. Hybridization facilitates evolutionary rescue. Evol. Appl. 7, 1209–1217 (2014).
Agarbati, A., Canonico, L., Comitini, F. & Ciani, M. Reduction of sulfur compounds through genetic improvement of native saccharomyces cerevisiae useful for organic and sulfite-free wine. Foods 9, (2020).
Kessi-Pérez, E. et al. Generation of a non-transgenic genetically improved yeast strain for wine production from nitrogen-deficient musts. Microorganisms 8, 1194 (2020).
Eberlein, C. et al. Hybridization is a recurrent evolutionary stimulus in wild yeast speciation. Nat. Commun. 10, (2019).
Bernardes, J. P., Stelkens, R. B. & Greig, D. Heterosis in hybrids within and between yeast species. J. Evol. Biol. 30, 538–548 (2017).
Banderas, A., Koltai, M., Anders, A. & Sourjik, V. Sensory input attenuation allows predictive sexual response in yeast. Nat. Commun. 7, 1–9 (2016).
Toyomura, K. & Hisatomi, T. Postzygotic reproductive isolation among three Saccharomyces yeast species. Yeast 38, 326–335 (2021).
Selmecki, A. M. et al. Polyploidy can drive rapid adaptation in yeast. Nature 519, 349–351 (2015).
Gerstein, A. C., Cleathero, L. A., Mandegar, M. A. & Otto, S. P. Haploids adapt faster than diploids across a range of environments. J. Evol. Biol. 24, 531–540 (2011).
Gerstein, A. C. & Otto, S. P. Cryptic fitness advantage: Diploids invade haploid populations despite lacking any apparent advantage as measured by standard fitness assays. PLoS One 6, (2011).
Bessho, K., Iwasa, Y. & Day, T. The evolutionary advantage of haploid versus diploid microbes in nutrient-poor environments. J. Theor. Biol. 383, 116–129 (2015).
Mable, B. K. Ploidy evolution in the yeast Saccharomyces cerevisiae: A test of the nutrient limitation hypothesis. J. Evol. Biol. 14, 157–170 (2001).
Horwitz, A. A. et al. Efficient multiplexed integration of synergistic alleles and metabolic pathways in yeasts via CRISPR-Cas. Cell Syst. 1, 88–96 (2015).
Gietz, R. D. & Schiestl, R. H. Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 35–37 (2007).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
Garrison, E. & Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 1–9 (2012).
Robinson, J. T. et al. Integrative genome viewer. Nat. Biotechnol. 29, 24–26 (2011).
Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Shafin, K. et al. Haplotype-aware variant calling with PEPPER-Margin-DeepVariant enables high accuracy in nanopore long-reads. Nat. Methods 18, 1322–1332 (2021).
Kolmogorov, M., Yuan, J., Lin, Y. & Pevzner, P. A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546 (2019).
Marçais, G., Deblasio, D. & Kingsford, C. Asymptotically optimal minimizers schemes. Bioinformatics 34, i13–i22 (2018).
O’Donnell, S. & Fischer, G. MUM&Co: Accurate detection of all SV types through whole-genome alignment. Bioinformatics 36, 3242–3243 (2020).
Hall, B. G., Acar, H., Nandipati, A. & Barlow, M. Growth rates made easy. Mol. Biol. Evol. 31, 232–238 (2014).
White, C. & Zainasheff, J. Yeast: The practical guide to beer fermentation. (Brewers Publications, 2010).
Nissen, T. L., Schulze, U., Nielsen, J. & Villadsen, J. Flux distributions in anaerobic, glucose-limited continuous cultures of saccharomyces cerevisiae. Microbiology 143, 203–218 (1997).
Krogerus, K., Holmström, S. & Gibson, B. Enhanced wort fermentation with de novo lager hybrids. Appl. Environ. Microbiol. 84, 1–20 (2018).
Steensels, J. et al. Improving industrial yeast strains: Exploiting natural and artificial diversity. FEMS Microbiol. Rev. 38, 947–995 (2014).
Zörgö, E. et al. Life history shapes trait heredity by accumulation of loss-of-function alleles in yeast. Mol. Biol. Evol. 29, 1781–1789 (2012).
Mardones, W. et al. Rapid selection response to ethanol in Saccharomyces eubayanus emulates the domestication process under brewing conditions. Microb. Biotechnol. 2, 1–18 (2021).
Acknowledgements
We thank Antonio Molina for technical help and Michael Handford (Universidad de Chile) for language support. We also acknowledge Fundación Ciencia & Vida for providing infrastructure, laboratory space and equipment for experiments.
Funding
This research was funded Agencia Nacional de Investigación y Desarrollo (ANID) FONDECYT program and ANID-Millennium Science Institute Program – ICN17_022. FC is supported by FONDECYT grant N° 1180161, FS by FONDECYT grant N° 1210955, JM by FONDECYT POSTDOCTORADO grant N° 3200545 and PV by ANID FONDECYT POSTDOCTORADO grant N° 3200575. RN is supported by FIC ‘Transferencia Levaduras Nativas para Cerveza Artesanal’ and FONDECYT grant N° 1180917. We also acknowledge Fundación Ciencia & Vida for providing infrastructure, laboratory space and equipment for experiments. This research was partially supported by the supercomputing infrastructure of the National Laboratory for High Performance Computing Chile (NLHPC, ECM-02).
Author information
Authors and Affiliations
Contributions
Conceptualization, J.M., F.S. and F.A.C.; methodology, J.M., C.V., F.S. and F.A.C.; software, C.I.O., P.V. and C.A.V.; validation, J.M., F.S., R.F.N. and F.A.C.; formal analysis, J.M., K.U., C.I.O. and C.A.V.; investigation, J.M., K.U., C.V., V.A., C.I.O. and C.A.V.; resources, J.M., F.S., R.F.N. and F.A.C.; data curation, C.I.O. and C.A.V.; writing—original draft preparation, J.M., C.A.V, P.V. and F.A.C.; writing—review and editing, J.M., K.U., C.V., V.A., P.V., C.I.O., C.A.V., F.S., R.F.N. and F.A.C.; visualization, J.M..; supervision, F.S., R.F.N. and F.A.C.; project administration, F.A.C.; funding acquisition, F.S., R.F.N. and F.A.C. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Molinet, J., Urbina, K., Villegas, C. et al. A Saccharomyces eubayanus haploid resource for research studies. Sci Rep 12, 5976 (2022). https://doi.org/10.1038/s41598-022-10048-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-10048-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.